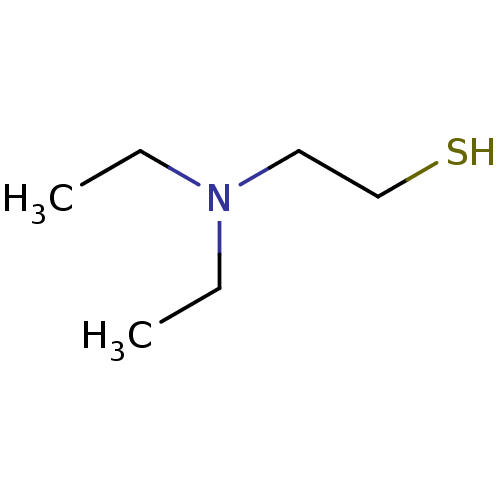
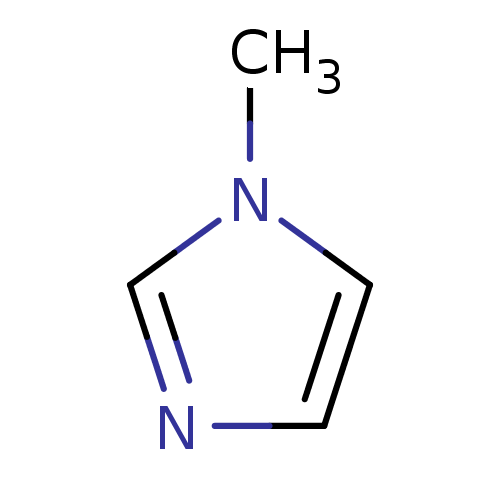
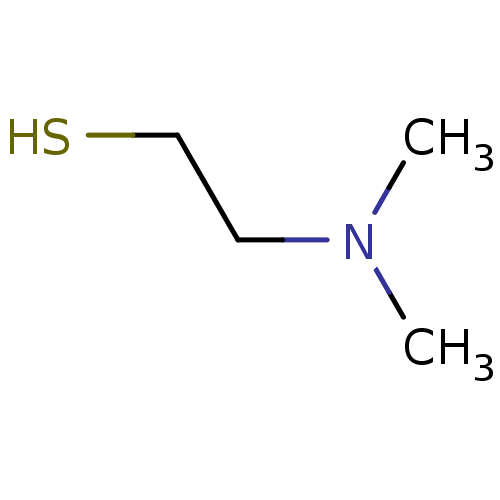
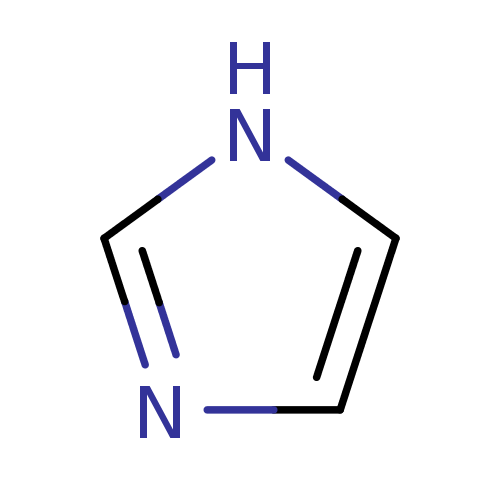
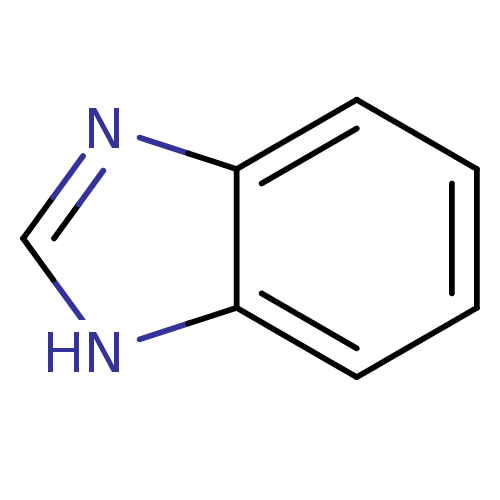
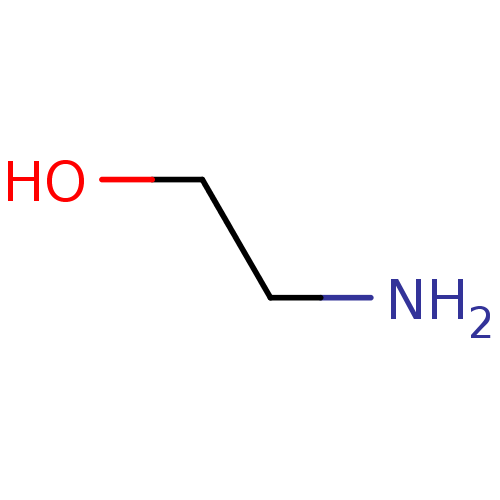
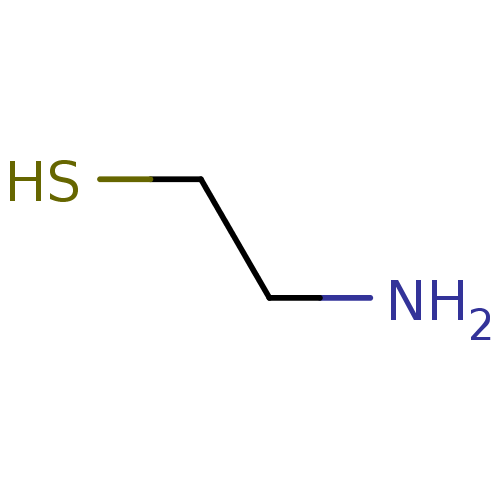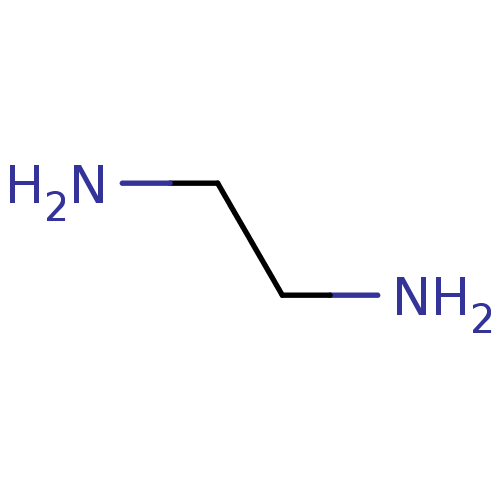Affinity DataIC50: 7.60nMAssay Description:Inhibition of mouse QPCT measured after 15 mins by fluorimetryMore data for this Ligand-Target Pair
Affinity DataIC50: 13nMAssay Description:Inhibition of mouse glutaminyl cyclase assessed as reduction in conversion of H-Gln-AMC hydrobromide to pGlu-AMC preincubated with substrate for 10 m...More data for this Ligand-Target Pair
Affinity DataIC50: 18nMAssay Description:Inhibition of mouse glutaminyl cyclaseMore data for this Ligand-Target Pair
Affinity DataIC50: 22nMAssay Description:Inhibition of mouse glutaminyl cyclase assessed as reduction in conversion of H-Gln-AMC hydrobromide to pGlu-AMC preincubated with substrate for 10 m...More data for this Ligand-Target Pair
Affinity DataIC50: 28nMAssay Description:Inhibition of mouse glutaminyl cyclaseMore data for this Ligand-Target Pair
Affinity DataIC50: 30nMAssay Description:Inhibition of mouse glutaminyl cyclaseMore data for this Ligand-Target Pair
Affinity DataIC50: 34nMAssay Description:Inhibition of mouse glutaminyl cyclaseMore data for this Ligand-Target Pair
Affinity DataIC50: 61nMAssay Description:Inhibition of mouse QPCT measured after 15 mins by fluorimetryMore data for this Ligand-Target Pair
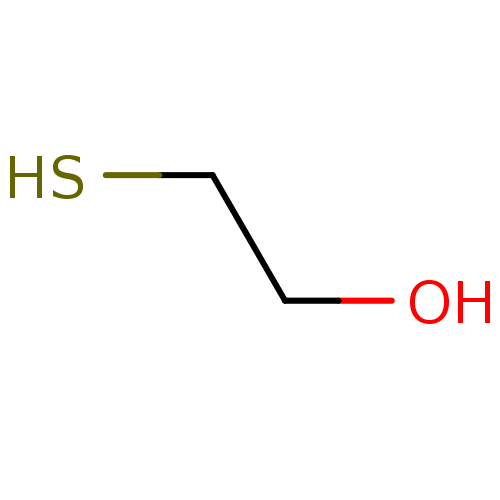
Affinity DataKi: 6.40E+3nM ΔG°: -30.1kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 1.09E+4nM ΔG°: -28.8kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 2.30E+4nM ΔG°: -26.9kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 2.90E+4nM ΔG°: -26.3kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 4.20E+4nM ΔG°: -25.4kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 1.60E+5nM ΔG°: -22.0kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 1.92E+5nM ΔG°: -21.6kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 1.93E+7nM ΔG°: -9.95kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 2.20E+7nM ΔG°: -9.62kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
Affinity DataKi: 2.58E+6nM ΔG°: -15.0kJ/molepH: 8.0 T: 2°CAssay Description:QC activity was evaluated fluorometrically using Gln-AMC as substrate, and pyroglutamyl peptidase as the auxiliary enzyme. After conversion of Gln-AM...More data for this Ligand-Target Pair
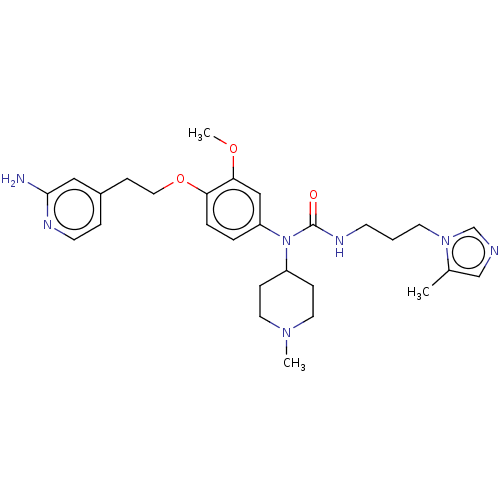


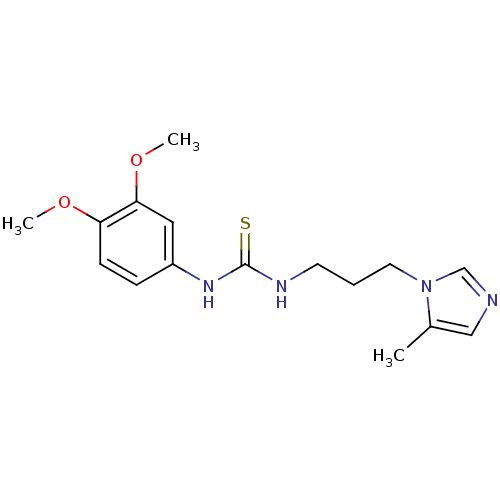
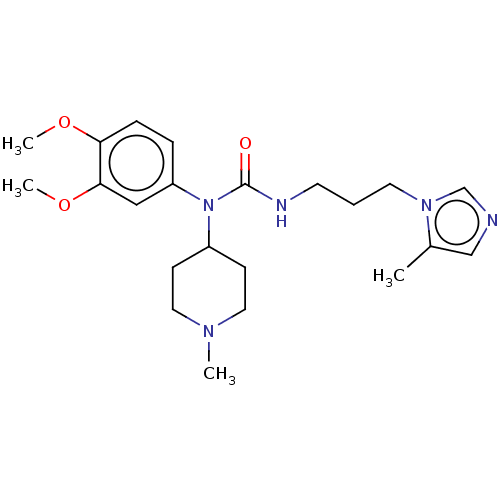
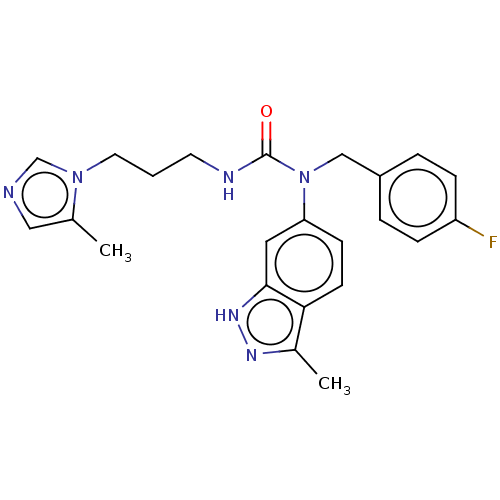
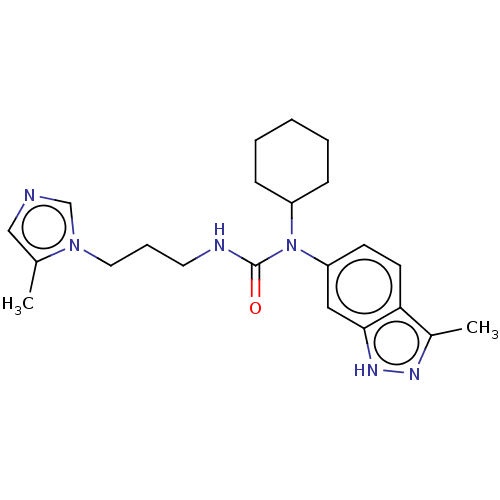
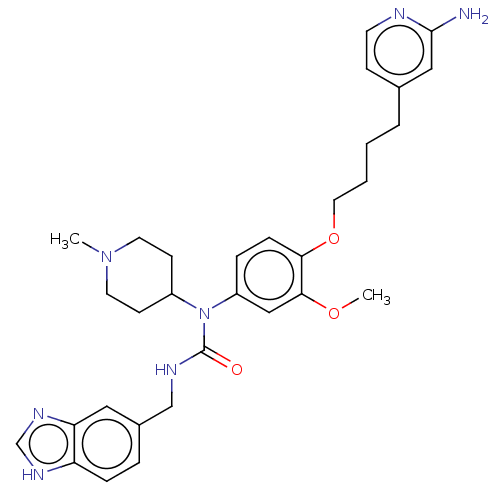
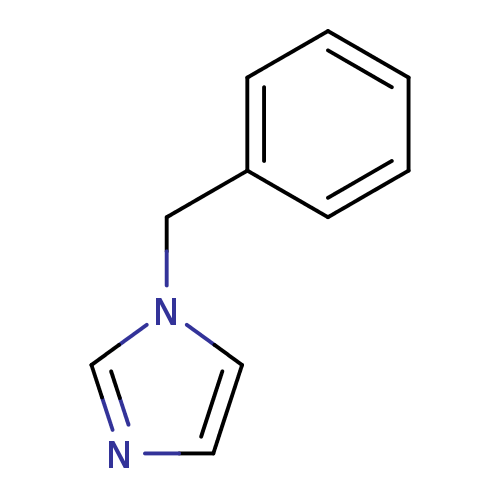
 3D Structure (crystal)
3D Structure (crystal)