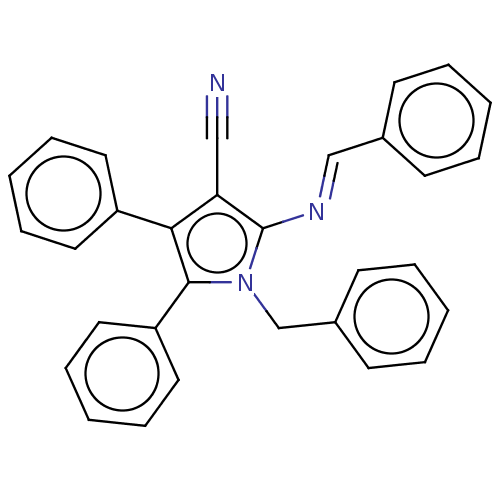
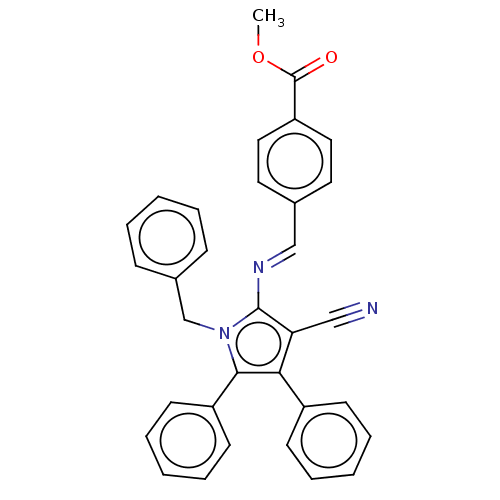
Report error Found 5 Enz. Inhib. hit(s) with all data for entry = 50020253
TargetMetallo-beta-lactamase type 2(Pseudomonas aeruginosa (g-Proteobacteria))
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.30E+3nMAssay Description:Competitive inhibition of NDM-1 (unknown origin) using penicillin G as substrate by Michaelis-Menten equation based kinetic analysisMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Pseudomonas aeruginosa (g-Proteobacteria))
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 1.70E+3nMAssay Description:Competitive inhibition of NDM-1 (unknown origin) using penicillin G as substrate by Michaelis-Menten equation based kinetic analysisMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Pseudomonas aeruginosa (g-Proteobacteria))
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 3.20E+3nMAssay Description:Competitive inhibition of NDM-1 (unknown origin) using penicillin G as substrate by Michaelis-Menten equation based kinetic analysisMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Pseudomonas aeruginosa (g-Proteobacteria))
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 9.00E+3nMAssay Description:Competitive inhibition of NDM-1 (unknown origin) using penicillin G as substrate by Michaelis-Menten equation based kinetic analysisMore data for this Ligand-Target Pair
TargetMetallo-beta-lactamase type 2(Pseudomonas aeruginosa (g-Proteobacteria))
The University of Queensland
Curated by ChEMBL
The University of Queensland
Curated by ChEMBL
Affinity DataKi: 3.40E+4nMAssay Description:Competitive inhibition of NDM-1 (unknown origin) using penicillin G as substrate by Michaelis-Menten equation based kinetic analysisMore data for this Ligand-Target Pair