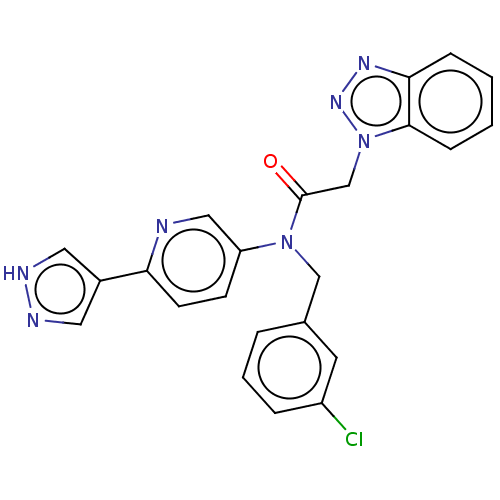
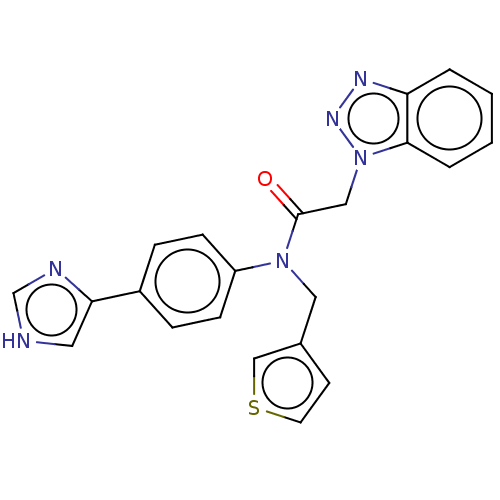
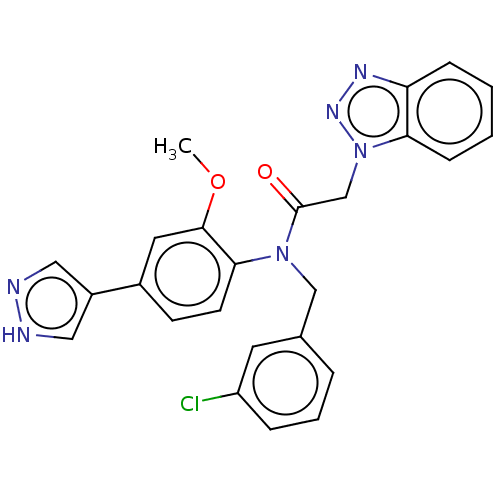
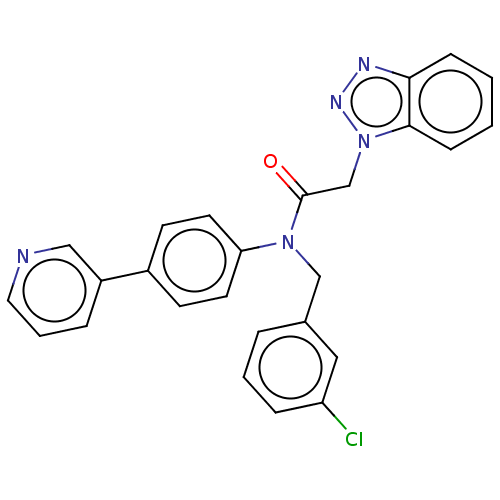
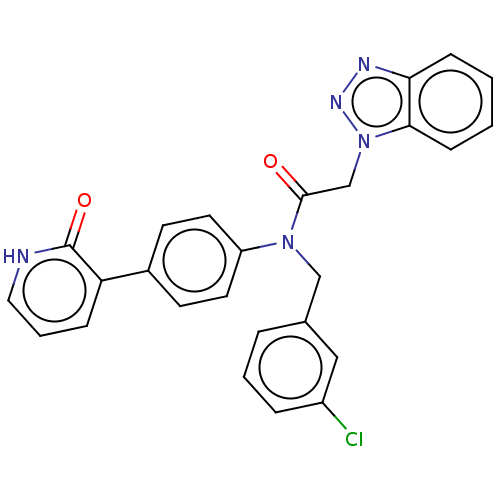
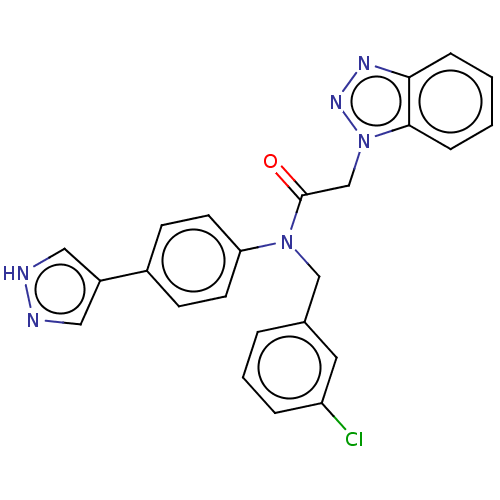
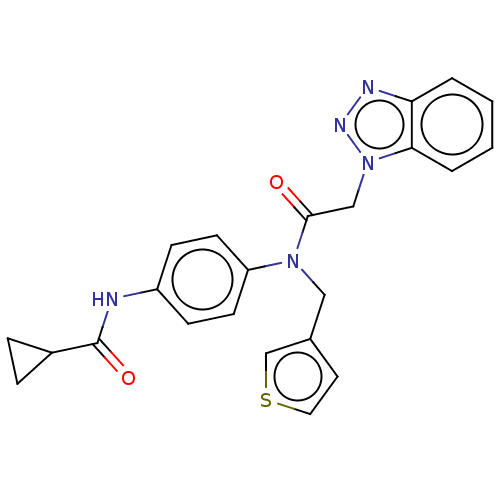
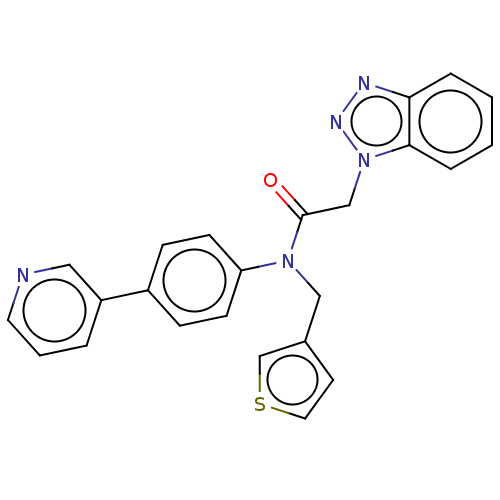
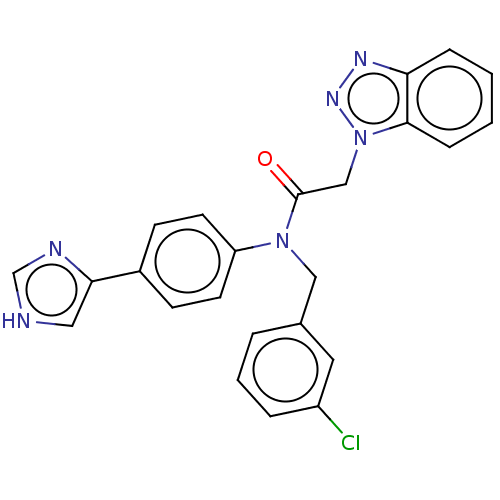
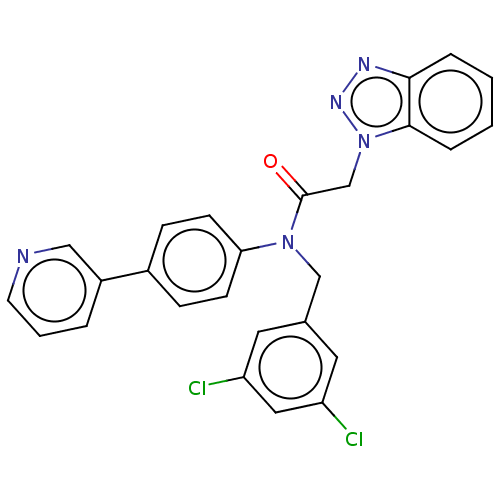
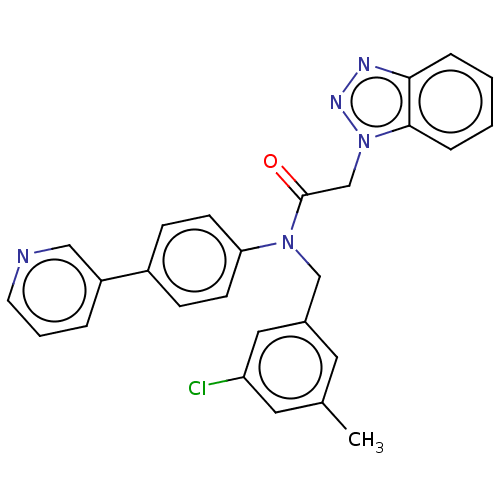
Report error Found 98 Enz. Inhib. hit(s) with all data for entry = 9660
Affinity DataEC50: 3.81E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 3.39E+3nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 1.91E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 5.76E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 7.63E+3nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 4.65E+3nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 1.74E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 1.75E+3nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 2.30E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 2.01E+3nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 7.61E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 1.11E+4nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 8.77E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 1.67E+4nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 1.98E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 3.31E+3nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 2.93E+3nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 3.24E+3nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 1.99E+4nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 2.82E+4nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 1.21E+4nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataEC50: 1.46E+4nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 560nMAssay Description:Confluent monolayers of Vero E6 ACE2 cells in 12-well plates were washed once with DMEM and infected with approximately 50 plaque forming units (PFUs...More data for this Ligand-Target Pair
Affinity DataEC50: 500nMAssay Description:Briefly, Vero E6 ACE2 cells were cultured in 96-well flat-bottom plates at a density of 2 × 104 cells per well. Following infection of the cells with...More data for this Ligand-Target Pair
Affinity DataIC50: 19nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 68nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 74nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 110nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 110nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 111nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 119nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 150nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 171nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 176nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 197nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 206nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 208nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 214nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 250nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 270nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 312nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 328nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 333nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 334nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 336nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 370nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 380nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 390nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair
Affinity DataIC50: 418nMAssay Description:The protease activity and subsequent 10-point IC50 curves were spectroscopically determined using a scaled down, end point assay adapted from a previ...More data for this Ligand-Target Pair