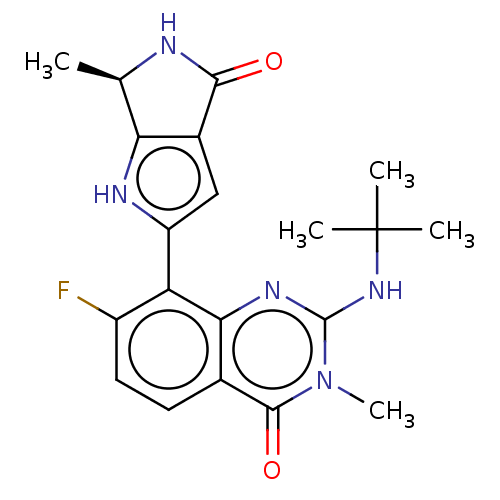
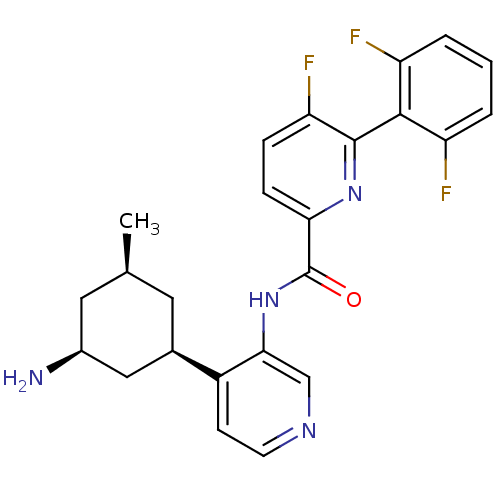
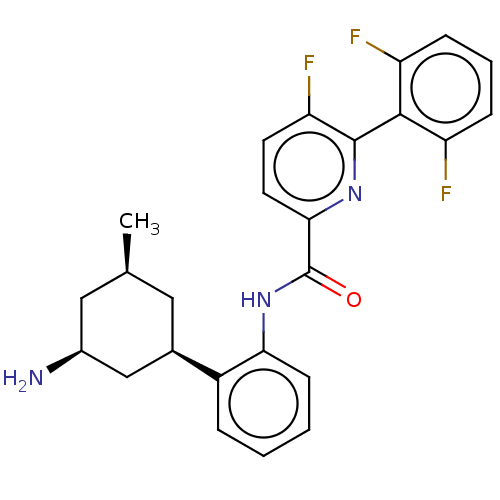
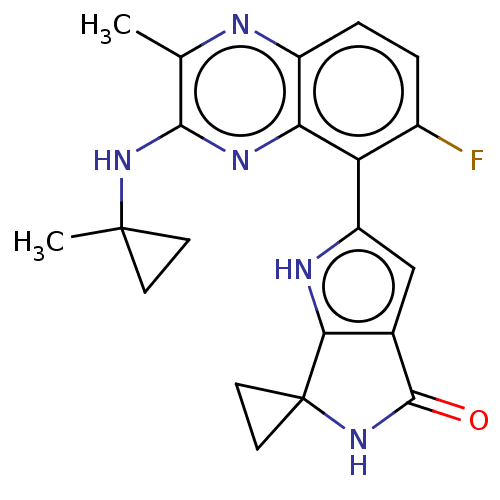
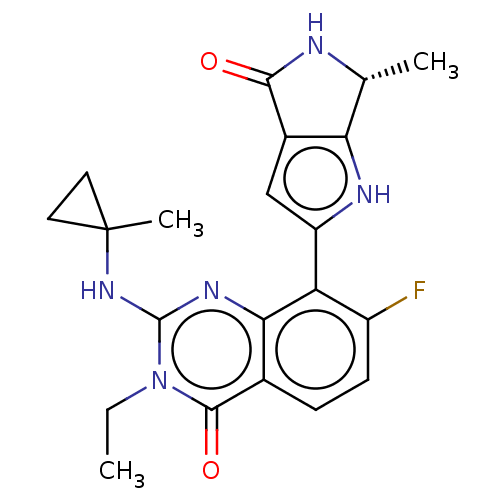
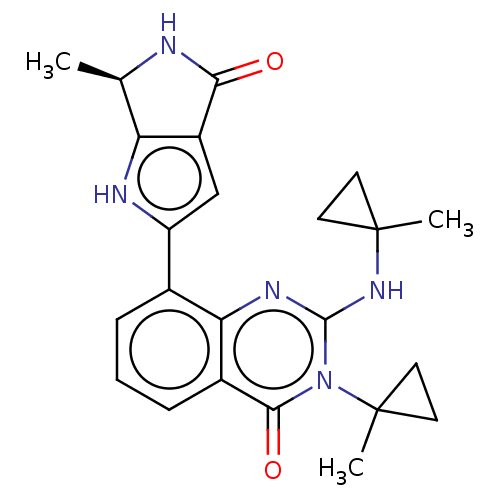
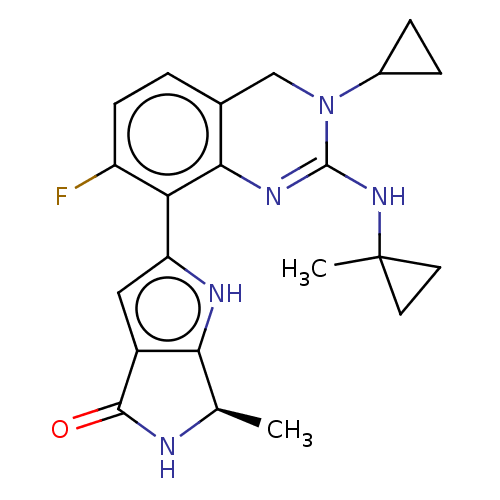
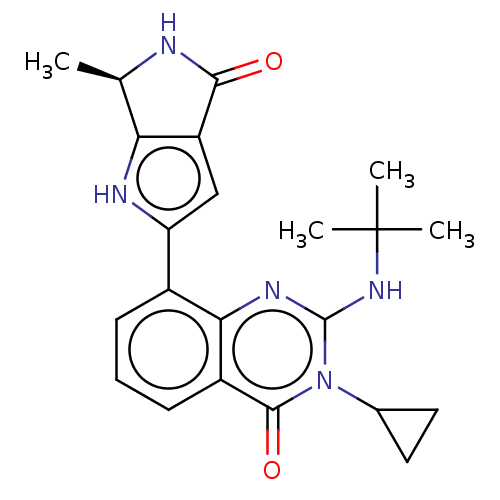
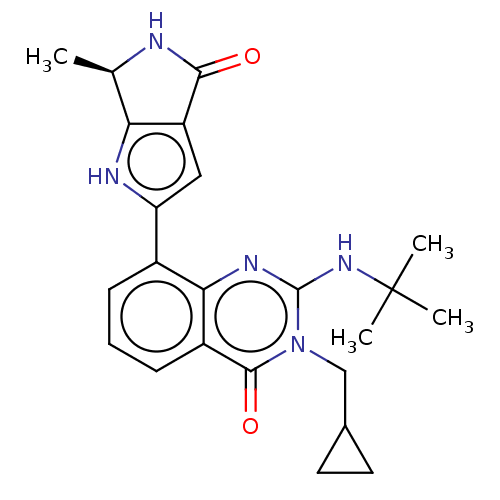
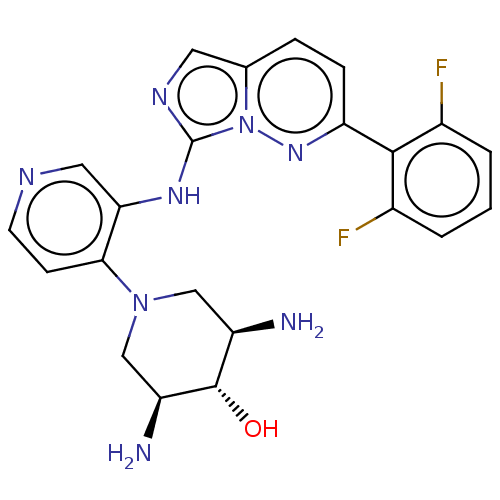
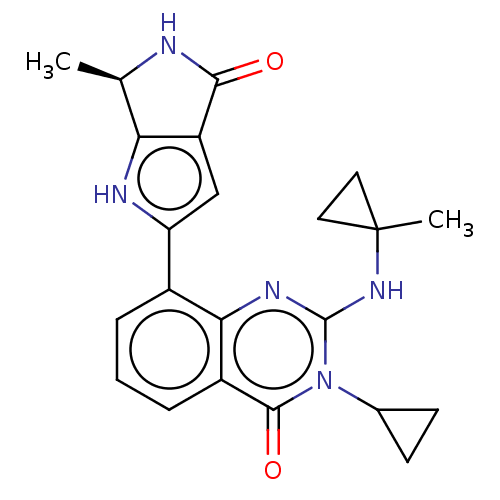
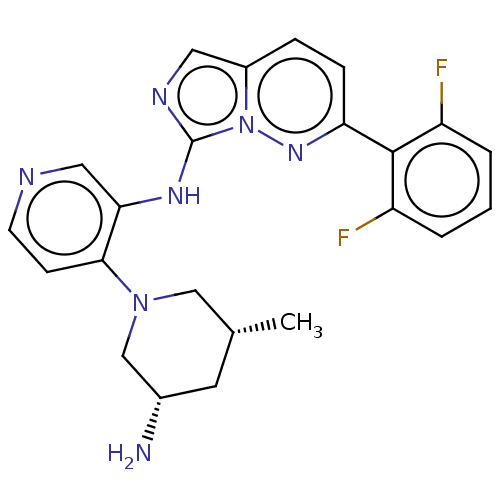
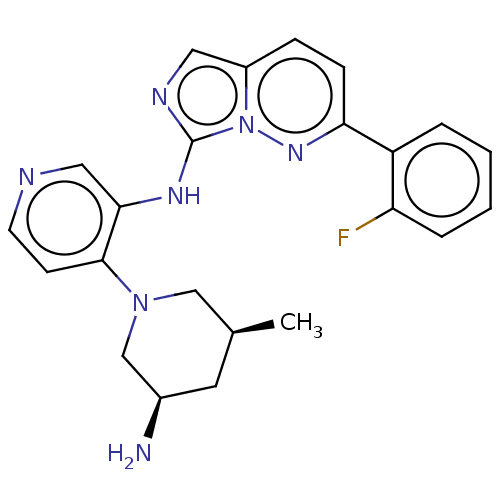
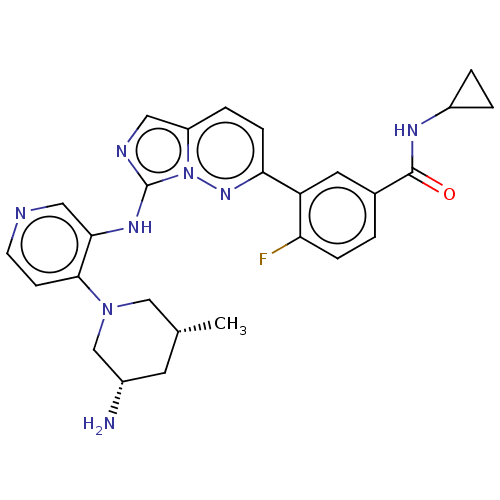
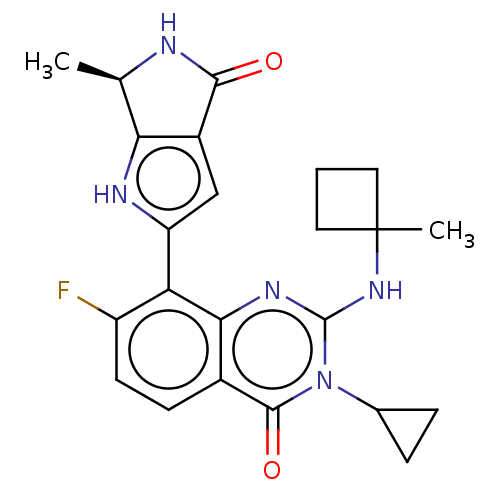
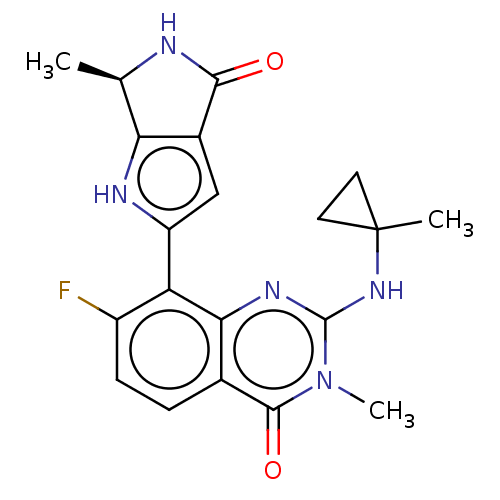
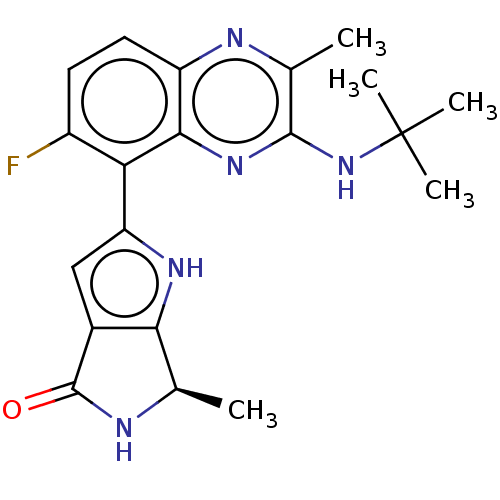
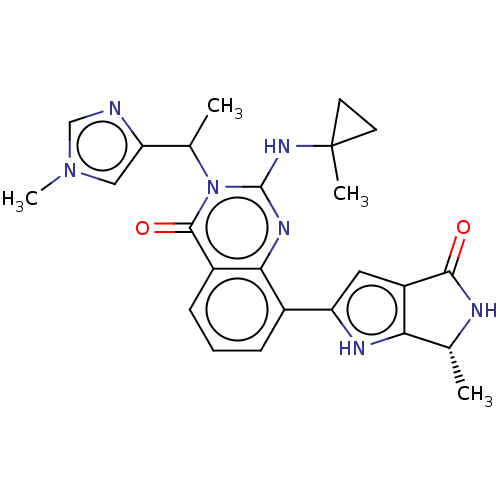
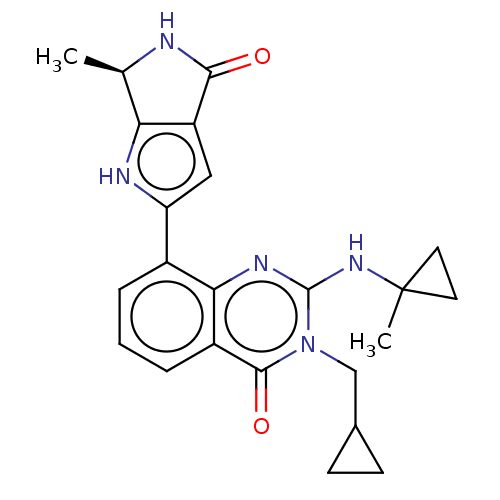
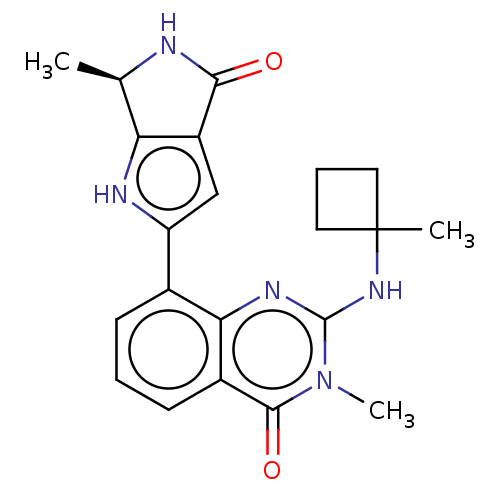
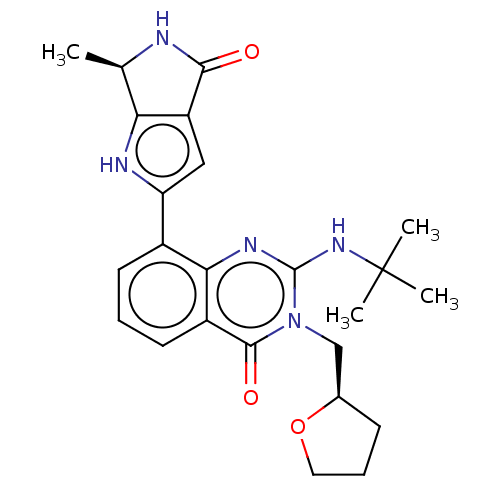
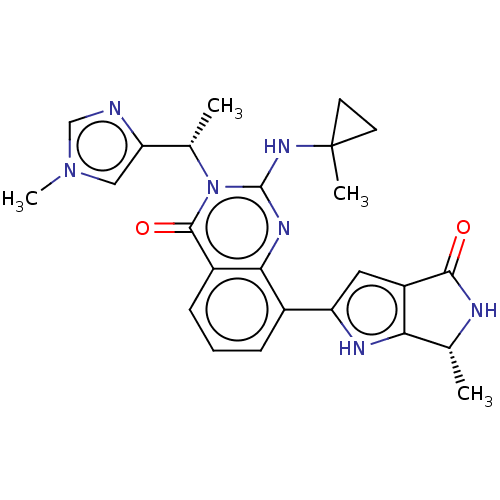
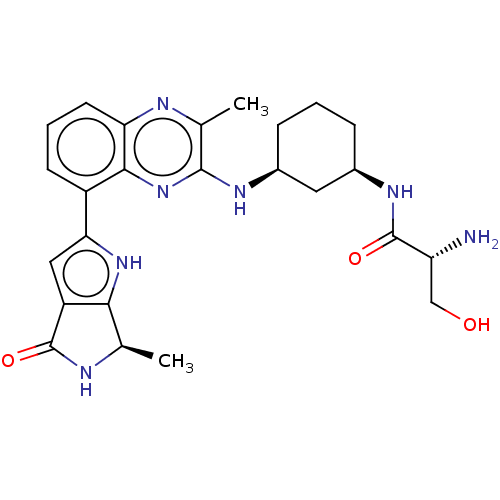
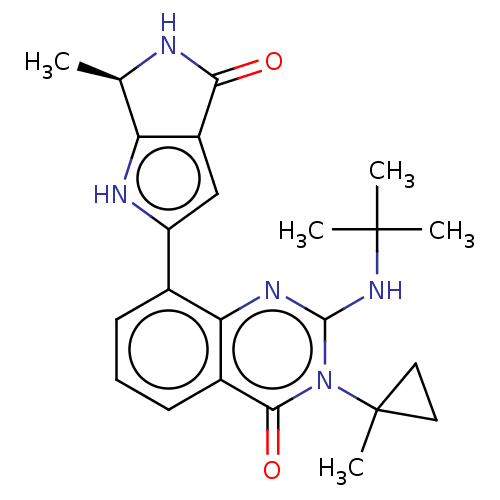
Report error Found 3843 of ic50 data for polymerid = 3422
Affinity DataIC50: 0.0120nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0120nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0180nMAssay Description:Inhibition of PIM2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.0180nMAssay Description:Inhibition of PIM2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 0.0200nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0354nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0358nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0378nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0378nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0484nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0500nMAssay Description:Inhibition of human full-length Pim2 expressed in Escherichia coli assessed as phosphorylation of biotinylated BAD peptide at Ser 112 preincubated fo...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0500nMAssay Description:Inhibition of human full-length Pim2 expressed in Escherichia coli assessed as phosphorylation of biotinylated BAD peptide at Ser 112 preincubated fo...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0500nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0600nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0667nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0700nMAssay Description:Inhibition of human full-length Pim2 expressed in Escherichia coli assessed as phosphorylation of biotinylated BAD peptide at Ser 112 preincubated fo...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0741nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0760nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0770nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0783nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0800nMAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0800nMAssay Description:Inhibition of human full-length Pim2 expressed in Escherichia coli assessed as phosphorylation of biotinylated BAD peptide at Ser 112 preincubated fo...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0800nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0900nMAssay Description:Inhibition of recombinant Pim2 (unknown origin) by electrochemiluminescence assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.0900nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.0940nMAssay Description:Inhibition of recombinant Pim2 (unknown origin) by electrochemiluminescence assayMore data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:Inhibition of full length recombinant Pim-2 (unknown origin) assessed as reduction in phosphorylation of biotinylated-BAD peptide at Serine 112 resid...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:Inhibition of full length recombinant Pim-2 (unknown origin) assessed as reduction in phosphorylation of biotinylated-BAD peptide at Serine 112 resid...More data for this Ligand-Target Pair
Affinity DataIC50: 0.100nMAssay Description:Inhibition of full length recombinant Pim-2 (unknown origin) assessed as reduction in phosphorylation of biotinylated-BAD peptide at Serine 112 resid...More data for this Ligand-Target Pair
Affinity DataIC50: 0.109nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.110nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.110nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.120nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.120nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.130nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.130nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.130nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.136nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.150nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair
Affinity DataIC50: 0.160nMpH: 7.0 T: 2°CAssay Description:The assay for the determination of Pim activity is based on the formation of phosphorylated biotinylated-BAD peptide at the Serine 112 residue (S112)...More data for this Ligand-Target Pair
Affinity DataIC50: 0.160nMAssay Description:Inhibition of full length recombinant PIM2 (unknown origin) assessed as reduction in BAD phosphorylation at Ser112 residues preincubated for 30 mins ...More data for this Ligand-Target Pair