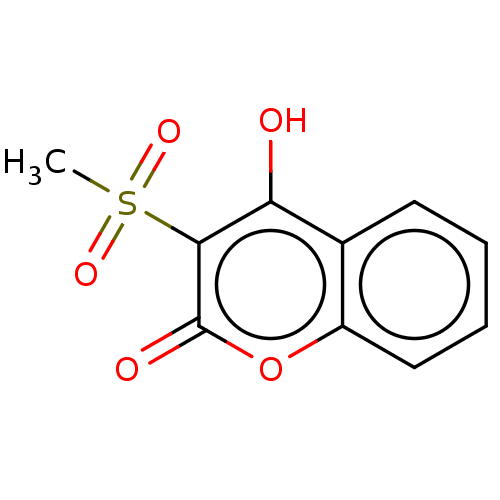
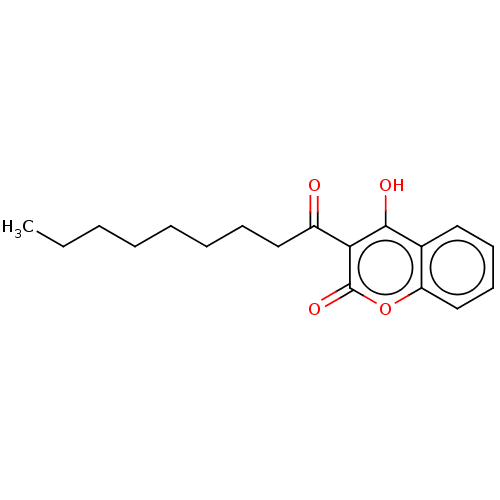
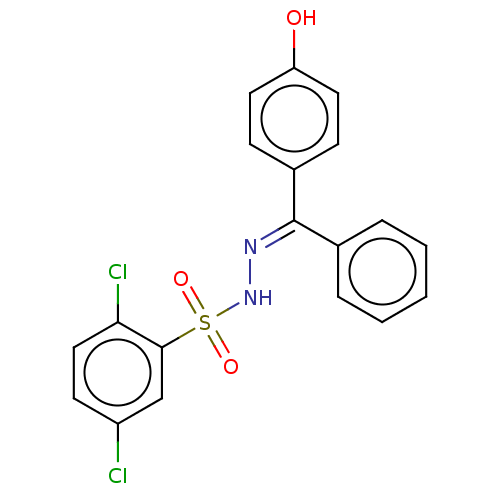
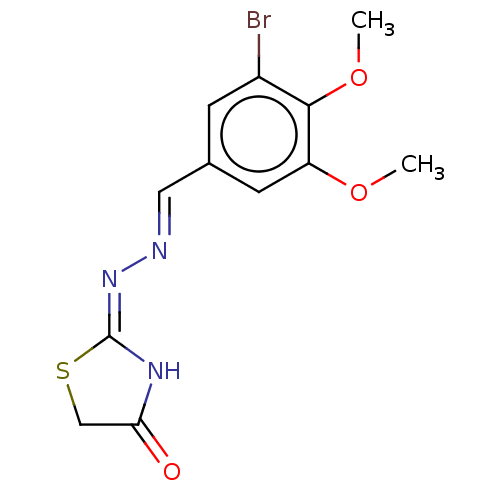
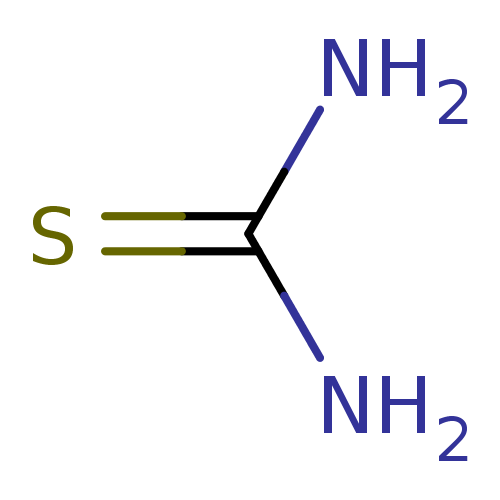Report error Found 94 Enz. Inhib. hit(s) with Target = 'Urease subunit alpha'
Displayed 1 to 50 (of 94 total ) | Next | Last >>

TargetUrease subunit alpha(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Jishou University
Curated by ChEMBL
Jishou University
Curated by ChEMBL
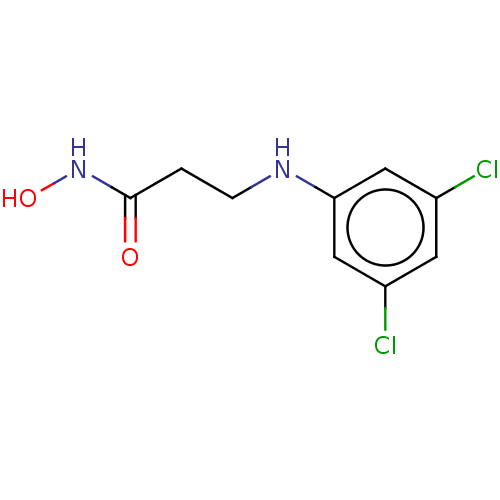
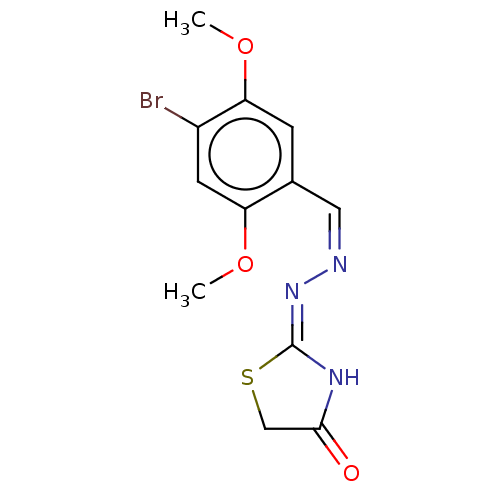
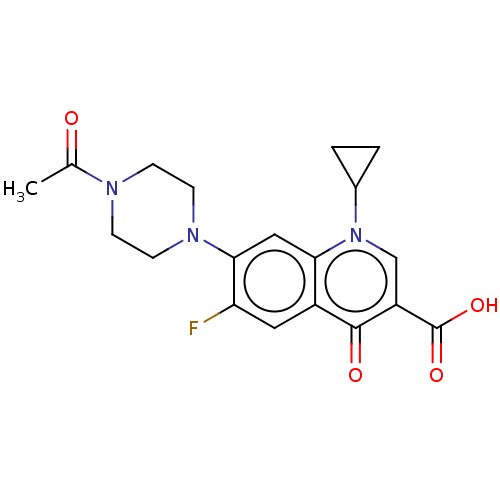
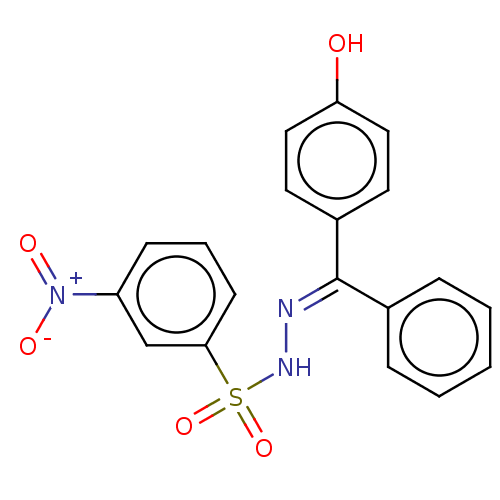
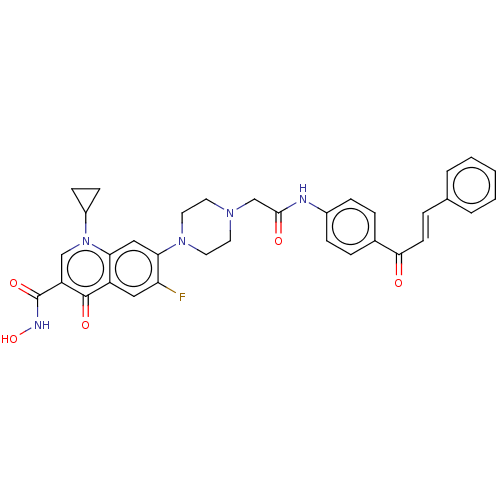
Affinity DataIC50: 43nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as reduction in ammonia production preincubated for 1.5 hrs under cell free condition by...More data for this Ligand-Target Pair
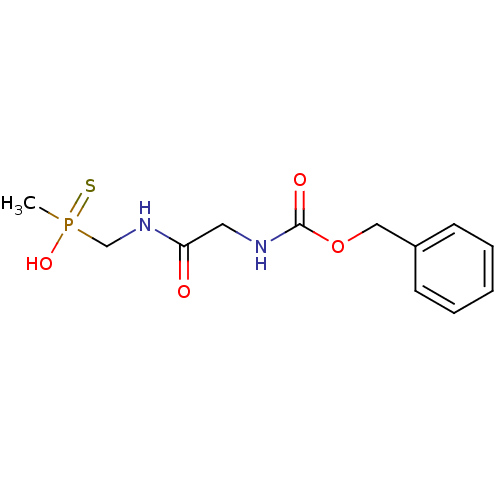
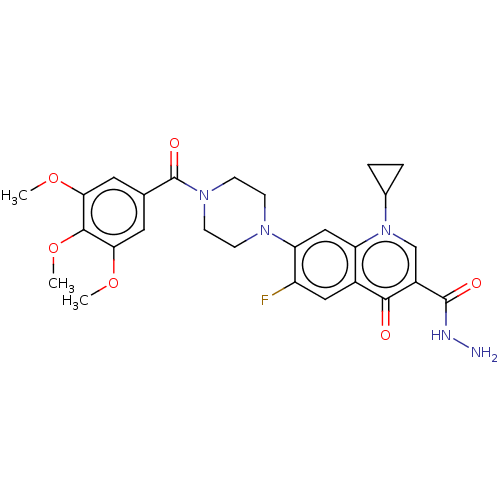
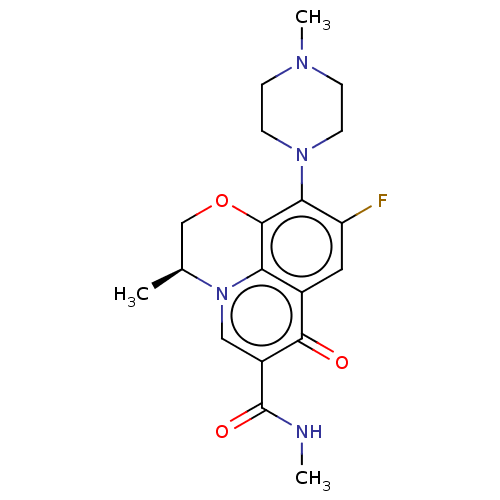
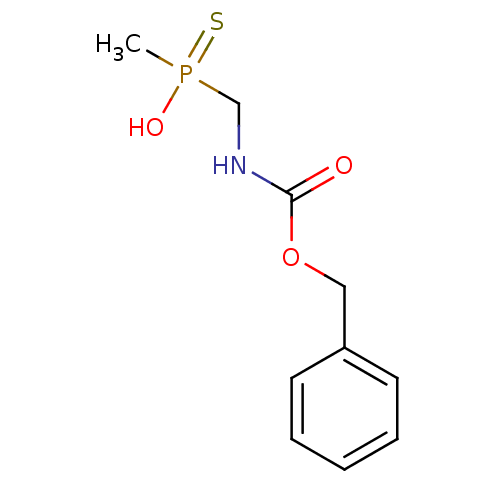
Affinity DataKi: 170nM ΔG°: -39.3kJ/mole IC50: 1.80E+3nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
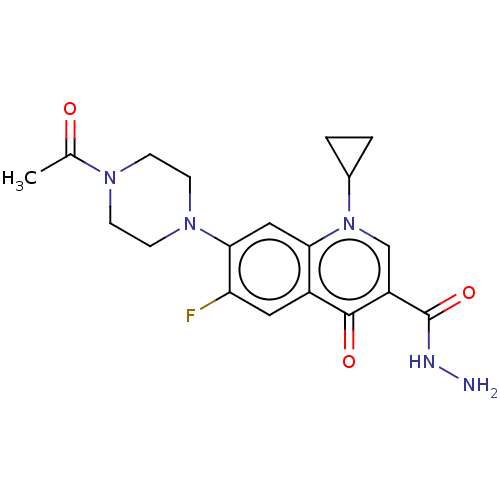
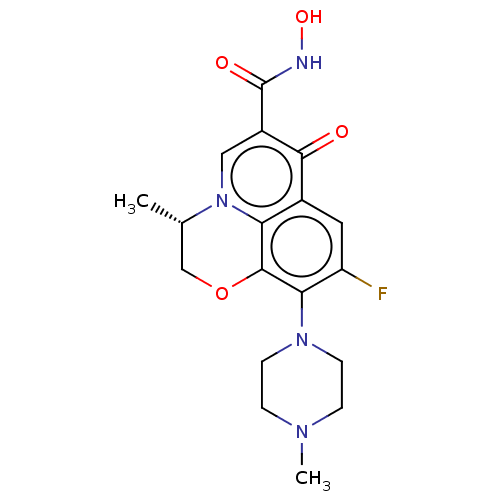
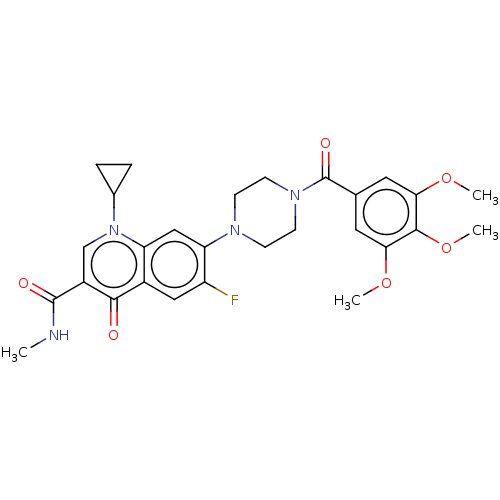
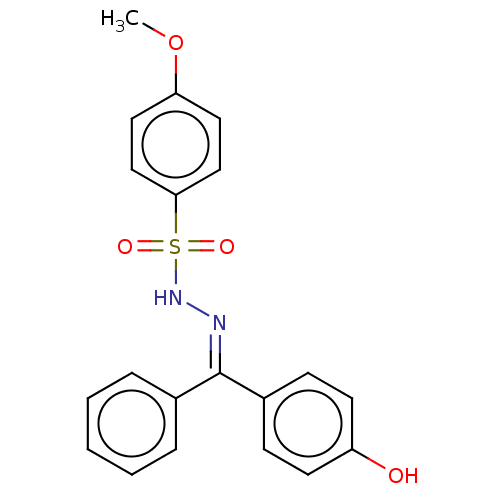
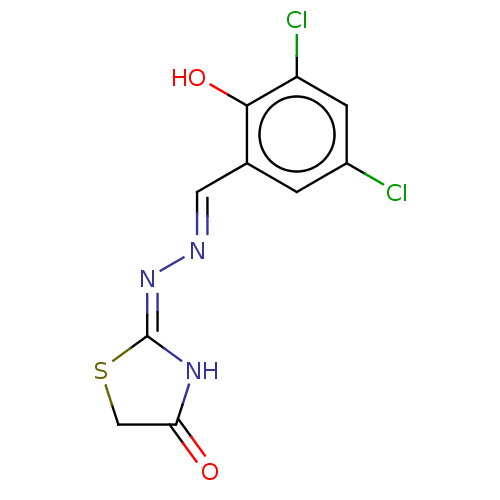
Affinity DataKi: 450nM ΔG°: -36.8kJ/mole IC50: 3.10E+3nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
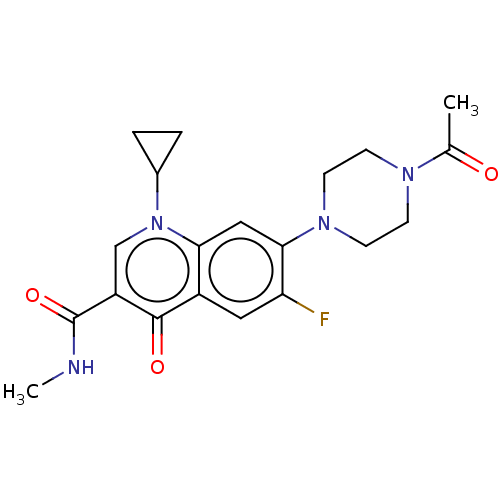
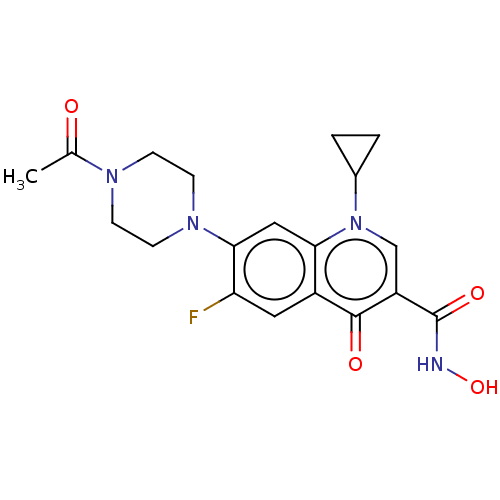
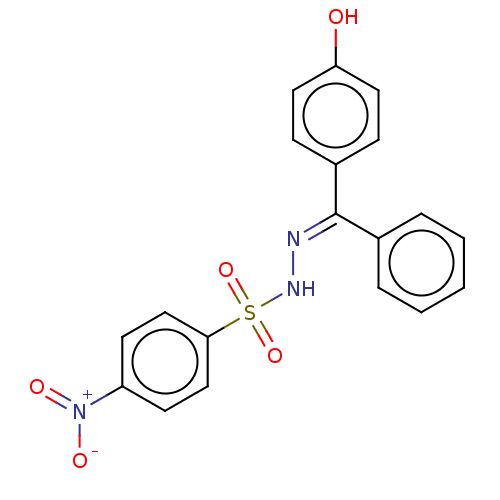
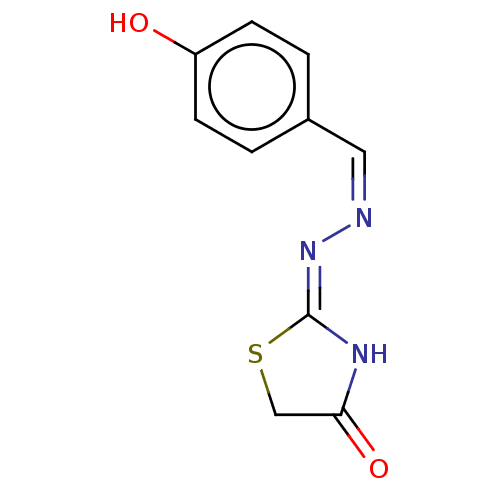
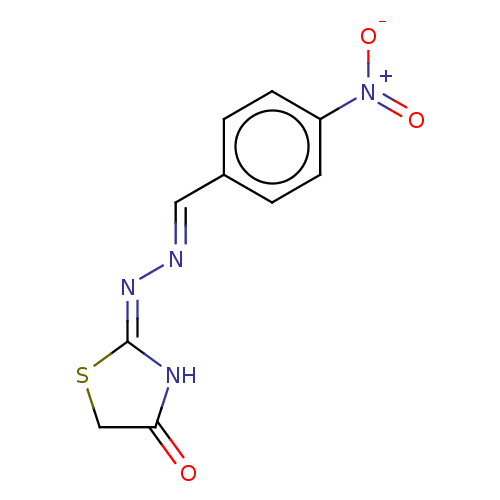
Affinity DataIC50: 1.22E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.29E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.73E+3nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.08E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.20E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.22E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.26E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.31E+3nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.89E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.92E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.90E+3nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 4.97E+3nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 5.70E+3nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 5.75E+3nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.17E+3nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 7.30E+3nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 8.43E+3nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 8.81E+3nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 9.34E+3nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.13E+4nMT: 2°CAssay Description:Reaction mixtures comprising one unit of urease enzyme (B. pasteurii) solution and 55 uL of buffers containing 100 mMol urea were incubated with 5 uL...More data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+4nMT: 2°CAssay Description:Reaction mixtures comprising one unit of urease enzyme (B. pasteurii) solution and 55 uL of buffers containing 100 mMol urea were incubated with 5 uL...More data for this Ligand-Target Pair
Affinity DataIC50: 1.37E+4nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataKi: 1.40E+4nM ΔG°: -28.2kJ/mole IC50: 1.58E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 1.45E+4nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 1.46E+4nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.63E+4nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 1.69E+4nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 1.71E+4nMT: 2°CAssay Description:Reaction mixtures comprising one unit of urease enzyme (B. pasteurii) solution and 55 uL of buffers containing 100 mMol urea were incubated with 5 uL...More data for this Ligand-Target Pair
Affinity DataKi: 1.75E+4nM ΔG°: -27.6kJ/mole IC50: 1.12E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+4nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataKi: 1.80E+4nM ΔG°: -27.5kJ/mole IC50: 6.00E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 1.89E+4nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataIC50: 1.93E+4nMAssay Description:The assay mixture, containing 50 μl (2 mg/ml) of enzyme and 100 μl of different concentration of the tested agents, was added to 850 μ...More data for this Ligand-Target Pair
Affinity DataKi: 2.10E+4nM ΔG°: -27.1kJ/mole IC50: 6.00E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 2.10E+4nMT: 2°CAssay Description:Reaction mixtures comprising one unit of urease enzyme (B. pasteurii) solution and 55 uL of buffers containing 100 mMol urea were incubated with 5 uL...More data for this Ligand-Target Pair
Affinity DataIC50: 2.13E+4nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataKi: 2.50E+4nM ΔG°: -26.7kJ/mole IC50: 8.00E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 2.67E+4nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataIC50: 2.69E+4nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair
Affinity DataKi: 2.70E+4nM ΔG°: -26.5kJ/mole IC50: 1.53E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 2.92E+4nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataIC50: 2.95E+4nMT: 2°CAssay Description:Reaction mixtures comprising one unit of urease enzyme (B. pasteurii) solution and 55 uL of buffers containing 100 mMol urea were incubated with 5 uL...More data for this Ligand-Target Pair
Affinity DataKi: 3.00E+4nM ΔG°: -26.2kJ/mole IC50: 8.60E+4nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 3.09E+4nMT: 2°CAssay Description:Reaction mixtures comprising one unit of urease enzyme (B. pasteurii) solution and 55 uL of buffers containing 100 mMol urea were incubated with 5 uL...More data for this Ligand-Target Pair
Affinity DataIC50: 3.12E+4nMAssay Description:Inhibition of bacterial urease using urea as substrate preincubated for 15 mins followed by substrate addition by ELISAMore data for this Ligand-Target Pair
Affinity DataKi: 3.26E+4nM ΔG°: -26.0kJ/mole IC50: 1.83E+5nMpH: 7.0 T: 2°CAssay Description:Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati...More data for this Ligand-Target Pair
Affinity DataIC50: 3.66E+4nMpH: 7.0Assay Description:This assay was modified from Berthelot assay and was employed for the determination of urease activity. The assay is based on the hydrolysis of urea ...More data for this Ligand-Target Pair