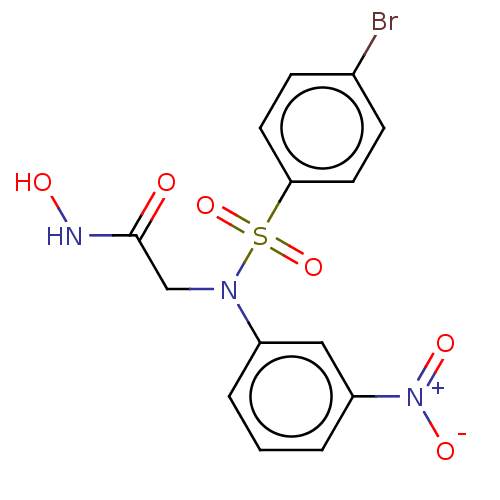
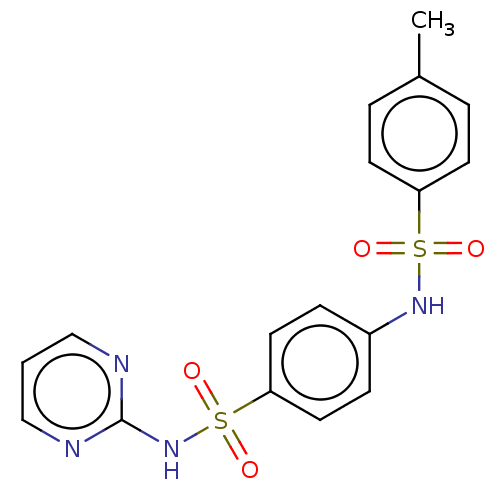
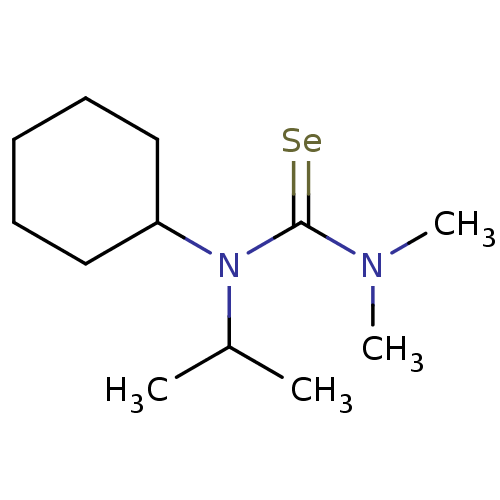
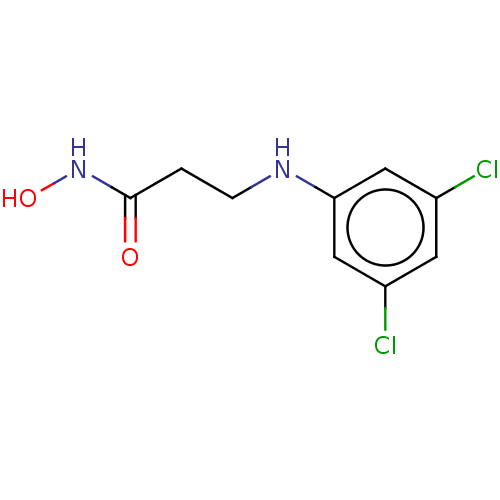
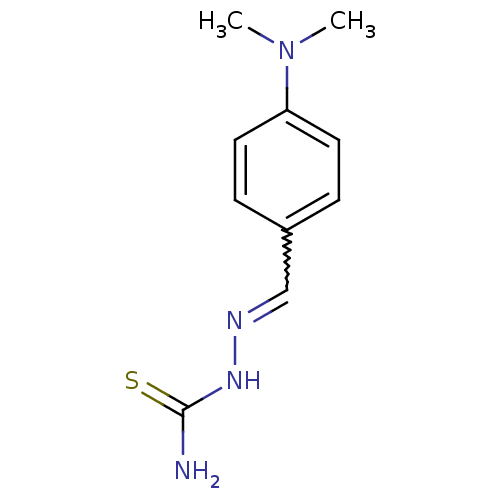
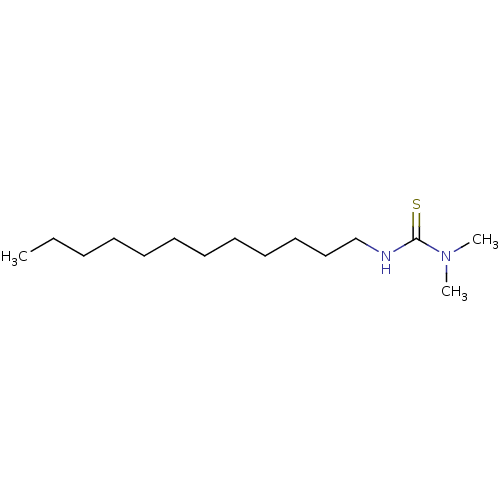
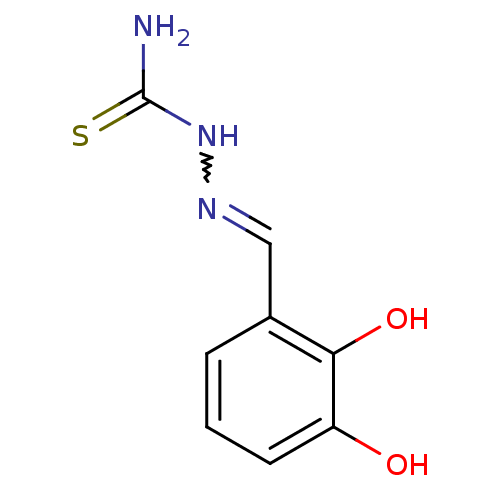
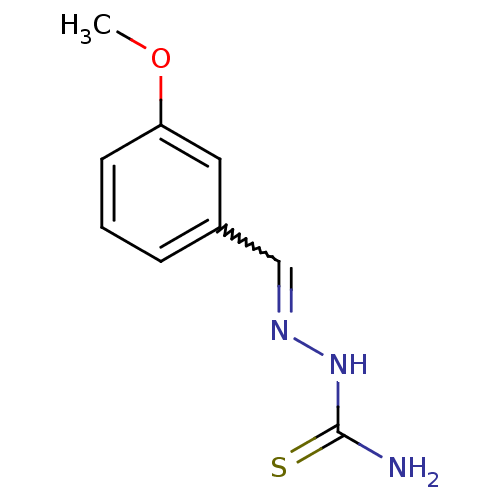
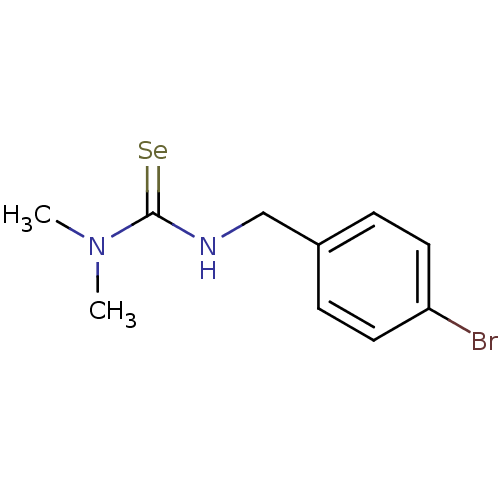
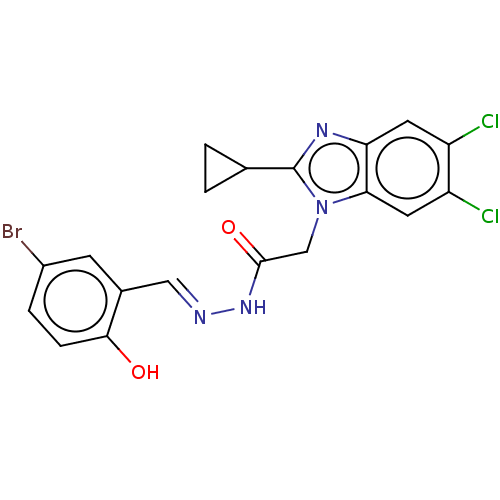
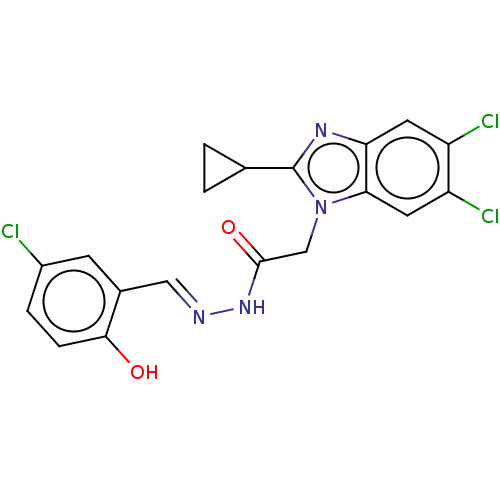
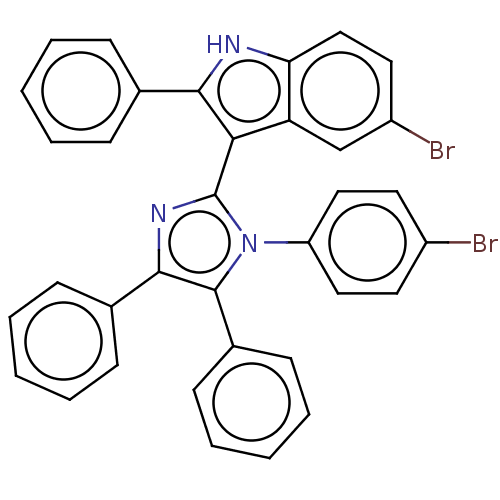
Report error Found 1162 Enz. Inhib. hit(s) with Target = 'Urease'
Displayed 1 to 50 (of 1162 total ) | Next | Last >>

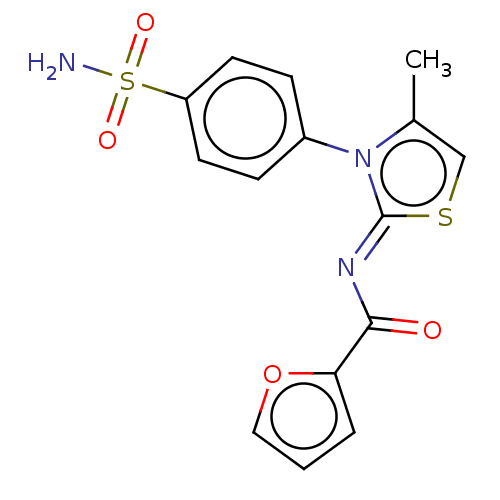
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
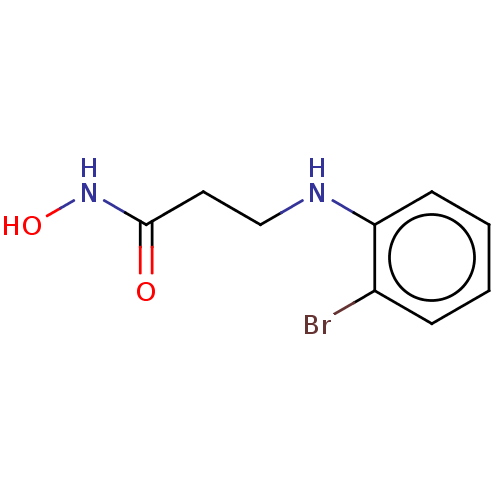
Affinity DataKd: 0.340nMAssay Description:Binding affinity to Helicobacter pyroli urease assessed as dissociation constant by surface plasmon resonance assayMore data for this Ligand-Target Pair
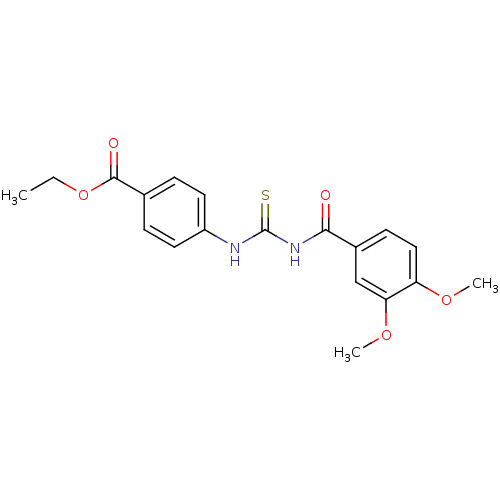
Affinity DataIC50: 2.20nMAssay Description:Inhibition of jack bean urease assessed as reduction in ammonia production using urea as substrate after 15 mins by indophenol methodMore data for this Ligand-Target Pair
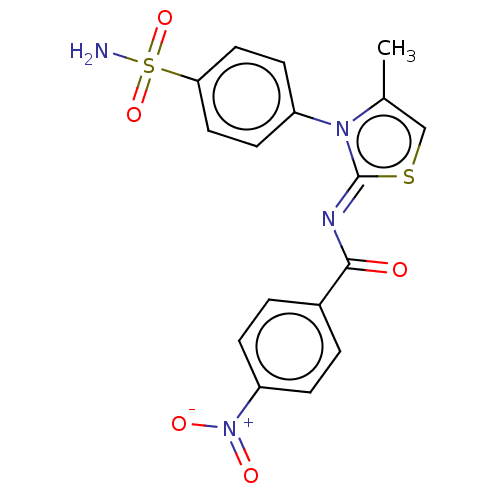
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
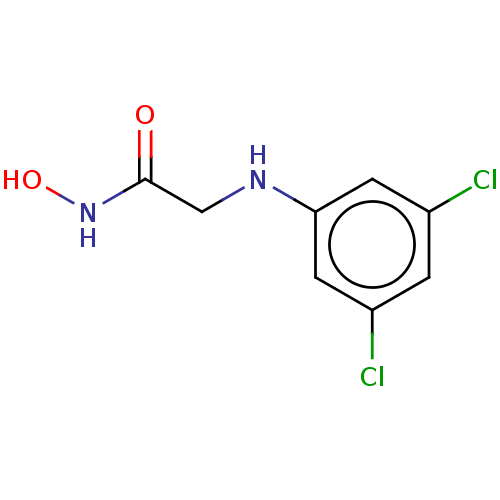
Affinity DataKi: 14nMAssay Description:Mixed-type competitive inhibition of Helicobacter pylori ATCC 43504 urease using urea as substrate by Lineweaver-burk plot analysisMore data for this Ligand-Target Pair
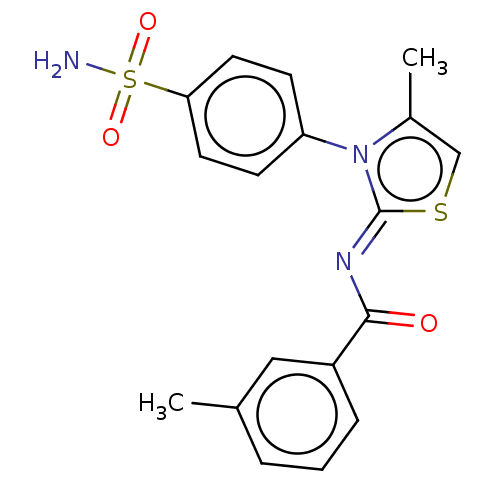
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
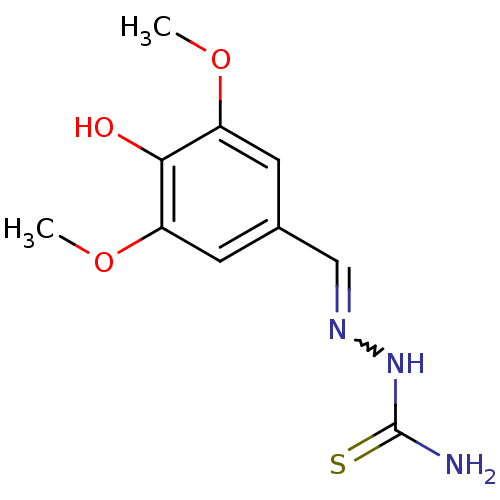
Affinity DataIC50: 18nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as reduction in ammonia production preincubated for 1.5 hrs under cell free condition by...More data for this Ligand-Target Pair
Affinity DataKi: 25nMAssay Description:Inhibition of jack bean ureaseMore data for this Ligand-Target Pair
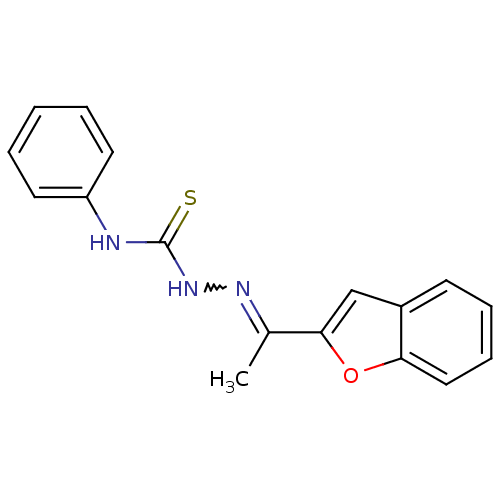
TargetUrease subunit alpha(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
Jishou University
Curated by ChEMBL
Jishou University
Curated by ChEMBL
Affinity DataIC50: 43nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as reduction in ammonia production preincubated for 1.5 hrs under cell free condition by...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataKi: 43nMAssay Description:Mixed-type inhibition of Helicobacter pylori ATCC 43504 urease assessed as enzyme-inhibitor complex using urea as substrate preincubated for 1.5 hrsMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataKi: 45nMAssay Description:Mixed-type competitive inhibition of Helicobacter pylori ATCC 43504 urease using urea as substrate by Lineweaver-burk plot analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 50nMAssay Description:Inhibition of jack bean urease using urea as substrate pretreated for 15 mins followed by substrate addition by phenol red based spectrophotometric m...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataKi: 54nMAssay Description:Mixed-type inhibition of Helicobacter pylori ATCC 43504 urease assessed as enzyme-substrate-inhibitor complex using urea as substrate preincubated fo...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 55nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as reduction in ammonia production preincubated for 1.5 hrs under cell free condition by...More data for this Ligand-Target Pair
Affinity DataIC50: 58nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:Inhibition of jack bean urease using urea as substrate pretreated for 15 mins followed by substrate addition by phenol red based spectrophotometric m...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataKi: 60.2nMAssay Description:Mixed type non-competitive inhibition of urease in Helicobacter pylori ATCC 43504 assessed as enzyme-substrate-inhibitor complex constant preincubate...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 61nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as reduction in ammonia production using urea as substrate preincubated for 1.5 hrs unde...More data for this Ligand-Target Pair
Affinity DataIC50: 64nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataKi: 65nMAssay Description:Mixed type inhibition of Helicobacter pyroli urease assessed as dissociation constant for EI complex by microplate reader analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 72nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 77nMpH: 8.2Assay Description:The mixture of this enzyme inhibition assay was composed of 10 µL of enzyme (5 U/mL) and 10 µL of each test compound in the series (3a-3s) in 40 µL b...More data for this Ligand-Target Pair
Affinity DataIC50: 81nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 83nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 83nMAssay Description:Inhibition of urease extracted from Helicobacter pylori ATCC 43504 assessed as ammonia production preincubated for 1.5 hrs by indophenol-based methodMore data for this Ligand-Target Pair
Affinity DataIC50: 83nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 87nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 87nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 89nMpH: 8.2Assay Description:The mixture of this enzyme inhibition assay was composed of 10 µL of enzyme (5 U/mL) and 10 µL of each test compound in the series (3a-3s) in 40 µL b...More data for this Ligand-Target Pair
Affinity DataKi: 90nMAssay Description:Inhibition of jack bean urease assessed as ammonia production after 30 mins by indophenol methodMore data for this Ligand-Target Pair
Affinity DataKi: 94nMAssay Description:Inhibition of jack bean ureaseMore data for this Ligand-Target Pair
Affinity DataIC50: 96nMpH: 8.2Assay Description:The urease inhibitory activity of synthesized sulfonamides (3a-j) was determined by measuring the amount of ammonia produced by the indophenols metho...More data for this Ligand-Target Pair
Affinity DataIC50: 102nMAssay Description:Inhibition of jack bean urease assessed as ammonia production after 30 mins by indophenol methodMore data for this Ligand-Target Pair
Affinity DataIC50: 102nMpH: 8.2Assay Description:The mixture of this enzyme inhibition assay was composed of 10 µL of enzyme (5 U/mL) and 10 µL of each test compound in the series (3a-3s) in 40 µL b...More data for this Ligand-Target Pair
Affinity DataIC50: 120nMAssay Description:Inhibition of jack beans urease assessed as hydrolysis of urea into ammonia preincubated for 10 mins followed by urea addition measured after 10 mins...More data for this Ligand-Target Pair
Affinity DataKi: 122nMAssay Description:Inhibition of jack bean urease assessed as ammonia production after 30 mins by indophenol methodMore data for this Ligand-Target Pair
Affinity DataKi: 124nMAssay Description:Inhibition of jack bean ureaseMore data for this Ligand-Target Pair
Affinity DataKi: 127nMAssay Description:Inhibition of jack bean urease assessed as ammonia production after 30 mins by indophenol methodMore data for this Ligand-Target Pair
Affinity DataIC50: 127nMAssay Description:Inhibition of jack bean urease assessed as ammonia production after 30 mins by indophenol methodMore data for this Ligand-Target Pair
Affinity DataKi: 127nMAssay Description:Inhibition of jack bean urease assessed as ammonia production after 30 mins by indophenol methodMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 130nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as reduction in ammonia production preincubated for 1.5 hrs under cell free condition by...More data for this Ligand-Target Pair
Affinity DataIC50: 130nMpH: 8.2 T: 2°CAssay Description:The assay mixture contained 40 µL of buffer (0.01 M LiCl2, 1 mM EDTA, 0.01M K2HPO4, pH 8.2) containing 100 mM urea, 10 µL of enzyme (5U/mL)...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 130nMAssay Description:Inhibition of Helicobacter pylori ureaseMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 130nMAssay Description:Inhibition of Helicobacter pylori ATCC 43504 urease assessed as reduction in ammonia production preincubated for 1.5 hrs under cell free condition by...More data for this Ligand-Target Pair
Affinity DataKi: 134nMAssay Description:Inhibition of jack bean urease assessed as ammonia production after 30 mins by indophenol methodMore data for this Ligand-Target Pair
Affinity DataKi: 135nMAssay Description:Inhibition of jack bean ureaseMore data for this Ligand-Target Pair
Affinity DataIC50: 140nMAssay Description:Inhibition of jack bean urease assessed as reduction in ammonia production using urea as substrate after 15 mins by indophenol methodMore data for this Ligand-Target Pair
Affinity DataIC50: 140nMAssay Description:Inhibition of jack bean urease using urea as substrate pretreated for 15 mins followed by substrate addition by phenol red based spectrophotometric m...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 140nMAssay Description:Inhibition of urease in Helicobacter pylori ATCC 43504 assessed as reduction in ammonia production preincubated for 1.5 hrs by indophenol methodMore data for this Ligand-Target Pair
Affinity DataIC50: 150nMAssay Description:Inhibition of jack bean urease using urea as substrate pretreated for 15 mins followed by substrate addition by phenol red based spectrophotometric m...More data for this Ligand-Target Pair
Affinity DataIC50: 150nMAssay Description:Inhibition of jack beans urease assessed as hydrolysis of urea into ammonia preincubated for 10 mins followed by urea addition measured after 10 mins...More data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataIC50: 150nMAssay Description:Inhibition of Helicobacter pylori urease using urea as substrate assessed as ammonia production after 50 mins by indophenol methodMore data for this Ligand-Target Pair
TargetUrease subunit beta(Helicobacter pylori (strain ATCC 700392 / 26695) (...)
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
National Demonstration Center For Experimental Chemistry Education
Curated by ChEMBL
Affinity DataKi: 152nMAssay Description:Mixed type non-competitive inhibition of urease in Helicobacter pylori ATCC 43504 assessed as enzyme-substrate-inhibitor complex constant preincubate...More data for this Ligand-Target Pair