Report error Found 40 Enz. Inhib. hit(s) with Target = 'Phosphoethanolamine N-methyltransferase'
Affinity DataKd: 800nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 2.20E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 2.90E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 5.80E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 1.07E+4nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 1.91E+4nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 4.72E+4nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 5.40E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 4.20E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 3.00E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 2.50E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 2.00E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 2.50E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 1.60E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 1.10E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 700nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 2.67E+4nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 1.66E+4nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 1.14E+4nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 8.20E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 6.10E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 1.30E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 900nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 8.60E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 6.00E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 9.30E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataKd: 6.10E+3nMpH: 7.5 T: 2°CAssay Description:For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm -mercaptoethanol, and 5% gly...More data for this Ligand-Target Pair
Affinity DataIC50: 1.25E+4nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
Affinity DataIC50: 3.38E+4nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
TargetPhosphoethanolamine N-methyltransferase(malaria parasite P. falciparum)
University of Siena
Curated by ChEMBL
University of Siena
Curated by ChEMBL
Affinity DataIC50: 4.43E+4nMAssay Description:Inhibition of plasmodium falciparum phosphoethanolamine N-methyltransferase expressed in Escherichia coli using phosphoethanolamine as substrate and ...More data for this Ligand-Target Pair
Affinity DataIC50: 9.15E+4nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
Affinity DataIC50: 1.25E+5nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
TargetPhosphoethanolamine N-methyltransferase(malaria parasite P. falciparum)
University of Siena
Curated by ChEMBL
University of Siena
Curated by ChEMBL
Affinity DataIC50: 4.60E+5nMAssay Description:Inhibition of Plasmodium falciparum phosphoethanolamine methyltransferase using phospethanolamine as substrate by radiochemical assay in presence of ...More data for this Ligand-Target Pair
TargetPhosphoethanolamine N-methyltransferase(malaria parasite P. falciparum)
University of Siena
Curated by ChEMBL
University of Siena
Curated by ChEMBL
Affinity DataIC50: 5.54E+5nMAssay Description:Inhibition of Plasmodium falciparum phosphoethanolamine methyltransferase using phospethanolamine as substrate by radiochemical assay in presence of ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.50E+6nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
Affinity DataIC50: 1.30E+7nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
Affinity DataIC50: 3.10E+6nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
Affinity DataIC50: 2.34E+7nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
Affinity DataIC50: 3.40E+6nMpH: 8.0 T: 2°CAssay Description:A radiochemical assay was used to measure enzymatic activity.More data for this Ligand-Target Pair
TargetPhosphoethanolamine N-methyltransferase(malaria parasite P. falciparum)
University of Siena
Curated by ChEMBL
University of Siena
Curated by ChEMBL
Affinity DataIC50: 1.35E+6nMAssay Description:Inhibition of Plasmodium falciparum phosphoethanolamine methyltransferase using phospethanolamine as substrate by radiochemical assay in presence of ...More data for this Ligand-Target Pair
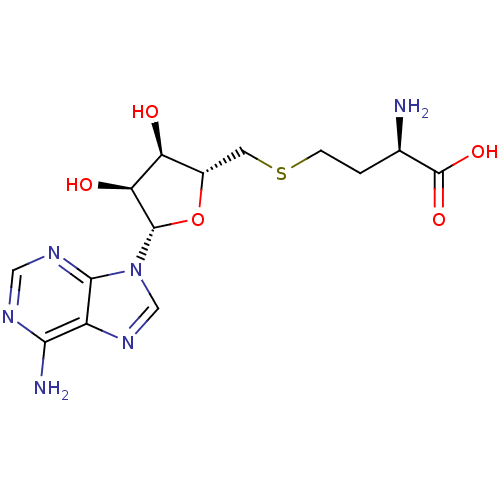
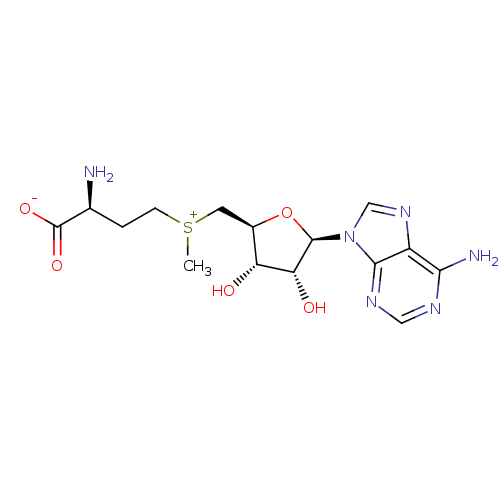
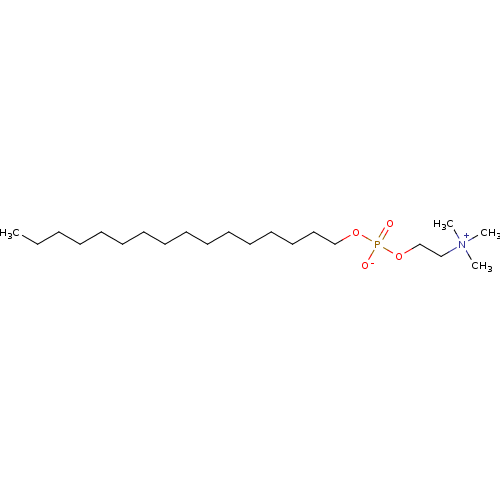
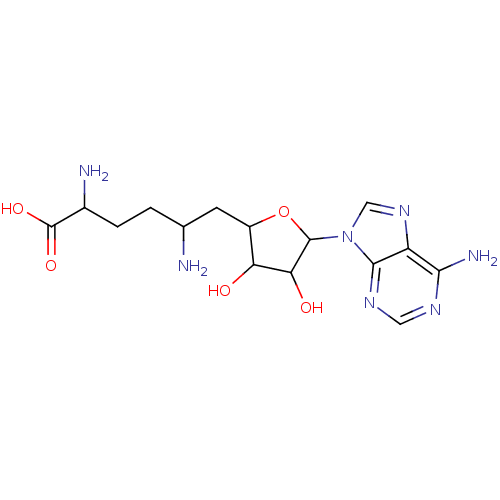
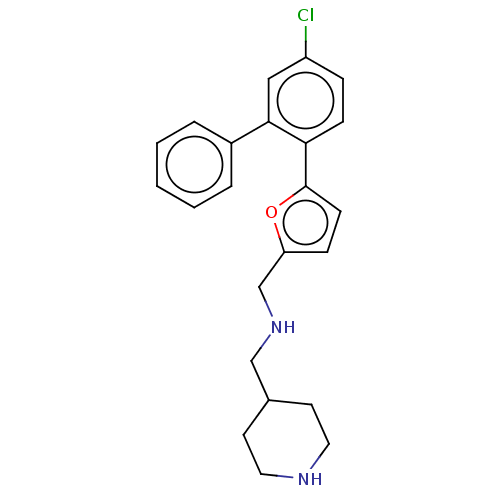

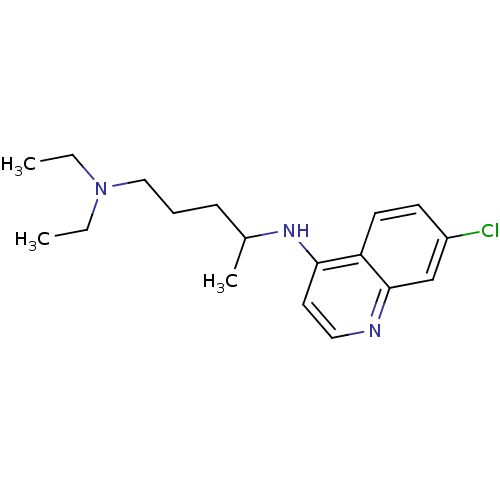


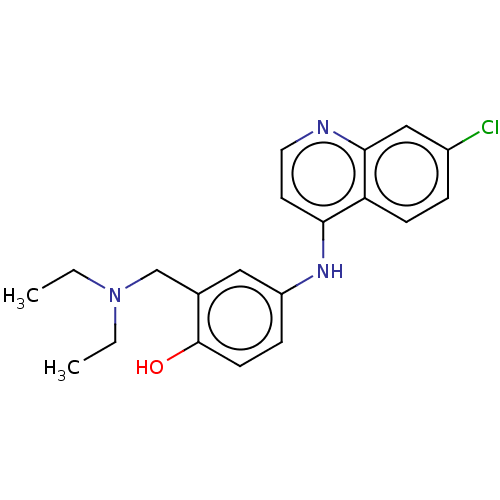
 3D Structure (crystal)
3D Structure (crystal)