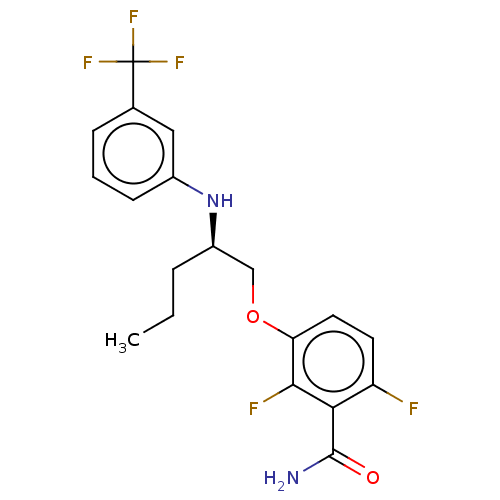
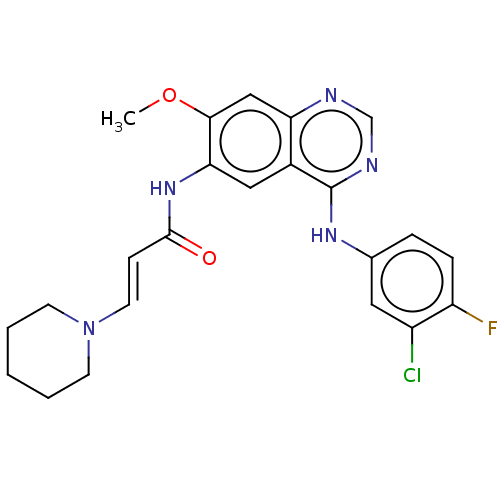
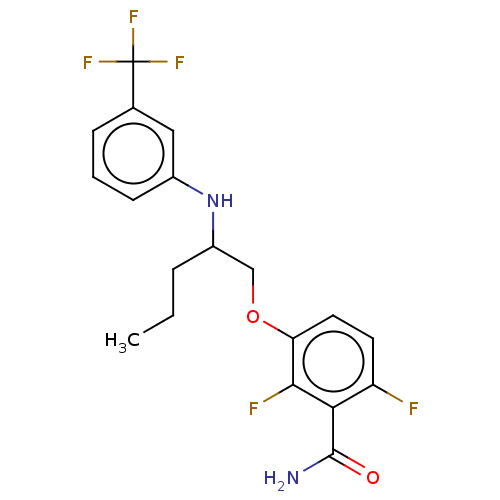
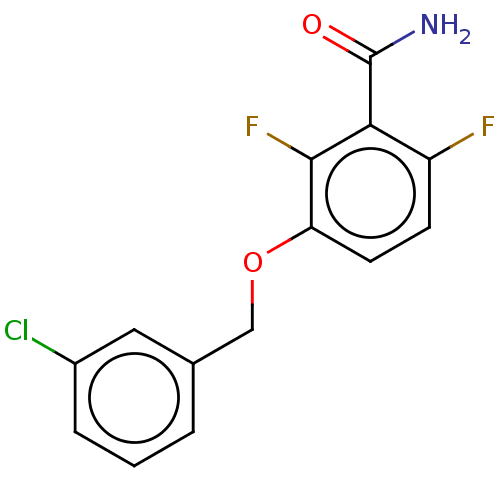
Report error Found 166 Enz. Inhib. hit(s) with Target = 'Cell division protein FtsZ'
Displayed 1 to 50 (of 166 total ) | Next | Last >>

Affinity DataKd: 3.03nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+4nM Kd: 3.33nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+4nM Kd: 15.6nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataKd: 23nMAssay Description:Displacement of bis-ANS from Escherichia coli FtsZMore data for this Ligand-Target Pair
Affinity DataIC50: 4.40E+4nM Kd: 71nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 3.50E+4nM Kd: 77nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataIC50: 4.10E+4nM Kd: 91nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataKd: <100nMAssay Description:Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.70E+4nM Kd: 126nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataIC50: 150nMAssay Description:Inhibition of Staphylococcus aureus FTsZ GTPase activityMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataIC50: 154nMAssay Description:Inhibition of Staphylococcus aureus FtsZ GTPase activityMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataIC50: 154nMAssay Description:Inhibition of Staphylococcus aureus FtsZ GTPase activityMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataIC50: 154nMAssay Description:Inhibition of Staphylococcus aureus FtsZ GTPase activityMore data for this Ligand-Target Pair
Affinity DataKd: 200nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 600nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
Affinity DataKd: 700nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1.39E+5nM Kd: 714nMpH: 7.5Assay Description:The binding affinities to FtsZ were determined by a fluorescent competition assay with 2'/3'-O-(N-methyl-anthraniloyl)-guanosine-5'-triph...More data for this Ligand-Target Pair
Affinity DataKd: 1.10E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.30E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Bacillus subtilis (strain 168))
Shandong University
Curated by ChEMBL
Shandong University
Curated by ChEMBL
Affinity DataKd: 1.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZMore data for this Ligand-Target Pair
Affinity DataKd: 1.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 1.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 2.00E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ expressed in Escherichia coli cells assessed as dissociation constant by fluorescence spectroscopyMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 2.38E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 2.49E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:Inhibition of FtsZ in Bacillus subtilis assessed as disruption of Z ring formationMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataIC50: 3.00E+3nMAssay Description:Inhibition of Escherichia coli recombinant FTsZ incubated for 10 mins in presence of GTP by spectrofluorimeter analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 3.50E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ expressed in Escherichia coli BL21 (DE3) at 25 degC by fluorescence anisotropy assayMore data for this Ligand-Target Pair
Affinity DataKd: 3.80E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataKd: 3.80E+3nMAssay Description:Binding affinity to Escherichia coli FtsZ at 25 degC by fluorescence anisotropy assayMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 3.84E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 3.86E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ assessed as dissociation constant by isothermal titration calorimetryMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataEC50: 4.00E+3nMAssay Description:Activation of Staphylococcus aureus FtsZ GTPase activityMore data for this Ligand-Target Pair
Affinity DataKd: 5.20E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ assessed as dissociation constant of intrinsic tryptophan quenching incubated for 30 mins by double recipr...More data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataKd: 5.40E+3nMAssay Description:Binding affinity to Staphylococcus aureus FtsZ expressed in Escherichia coli cells assessed as dissociation constant by fluorescence spectroscopyMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataIC50: 5.81E+3nMAssay Description:Inhibition of Escherichia coli FtsZ GTPase activityMore data for this Ligand-Target Pair
Affinity DataKd: 5.90E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
State University of New York
Curated by ChEMBL
State University of New York
Curated by ChEMBL
Affinity DataIC50: 6.21E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis FtsZ polymerizationMore data for this Ligand-Target Pair
Affinity DataKd: 6.70E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataIC50: 6.86E+3nMAssay Description:Inhibition of Escherichia coli FtsZ polymerizationMore data for this Ligand-Target Pair
Affinity DataKd: 7.10E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Mycobacterium tuberculosis (strain ATCC 25618 / H3...)
State University of New York
Curated by ChEMBL
State University of New York
Curated by ChEMBL
Affinity DataIC50: 7.69E+3nMAssay Description:Inhibition of Mycobacterium tuberculosis FtsZ polymerizationMore data for this Ligand-Target Pair
Affinity DataKd: 8.00E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataIC50: 8.30E+3nMAssay Description:Inhibition of Escherichia coli FtsZ GTPase activityMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataIC50: 9.00E+3nMAssay Description:Inhibition of Escherichia coli FtsZ GTPase activity after 5 mins by enzyme-coupled inhibition assayMore data for this Ligand-Target Pair
Affinity DataKd: 9.30E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
Affinity DataKd: 9.50E+3nMAssay Description:Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysisMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Escherichia coli (strain K12))
University of Wisconsin-Madison
Curated by ChEMBL
University of Wisconsin-Madison
Curated by ChEMBL
Affinity DataIC50: 9.87E+3nMAssay Description:Inhibition of Escherichia coli FtsZ GTPase activityMore data for this Ligand-Target Pair
TargetCell division protein FtsZ(Staphylococcus aureus)
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Indian Institute of Science Education and Research Bhopal
Curated by ChEMBL
Affinity DataIC50: 1.00E+4nMAssay Description:Inhibition of Staphylococcus aureus recombinant FTsZ preincubated for 10 mins followed by compound addition measured after 30 mins by Cytophos phosph...More data for this Ligand-Target Pair
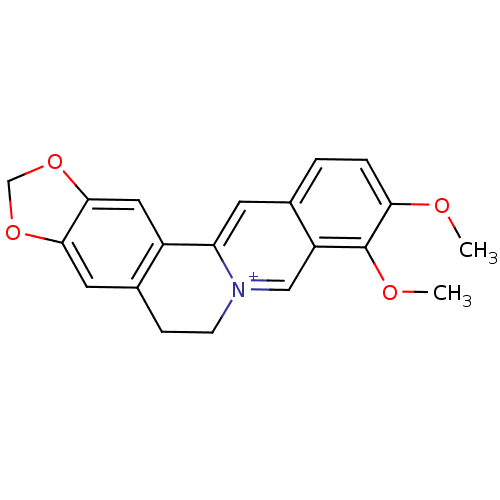
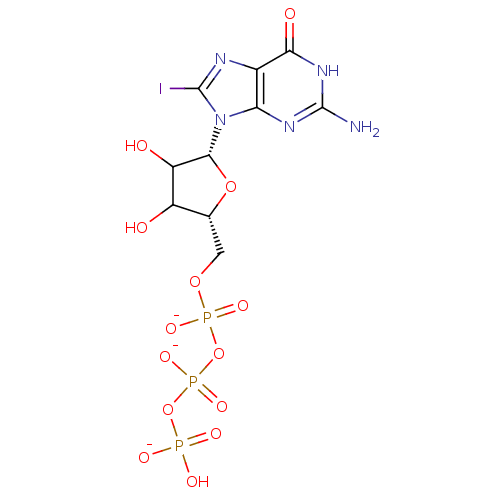
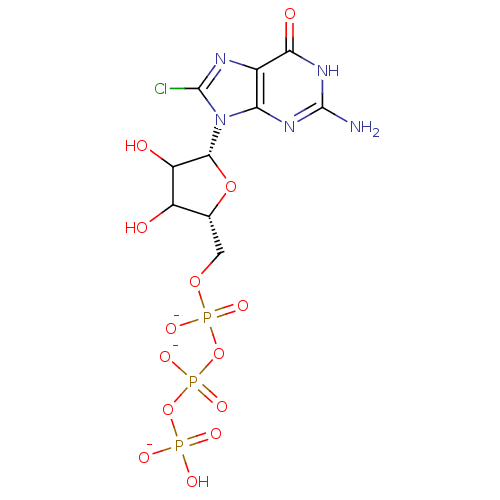
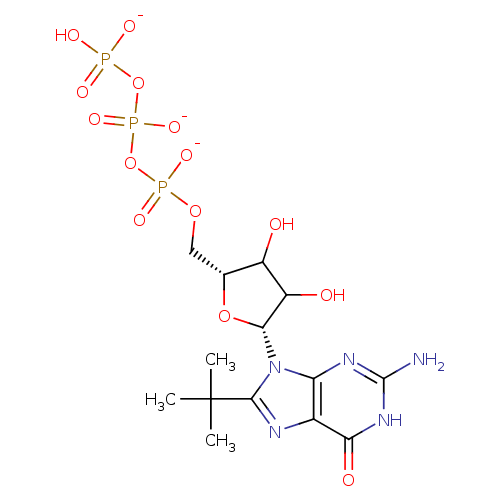
 3D Structure (crystal)
3D Structure (crystal)