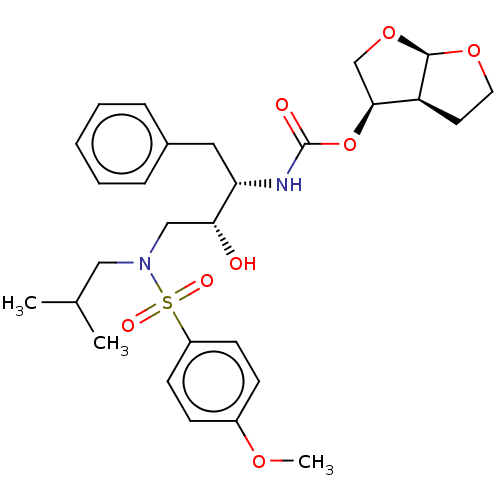
BDBM9236 (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S,3R)-3-hydroxy-4-[(4-methoxybenzene)(2-methylpropyl)sulfonamido]-1-phenylbutan-2-yl]carbamate::TMC-126::UIC-94003
SMILES COc1ccc(cc1)S(=O)(=O)N(CC(C)C)C[C@@H](O)[C@H](Cc1ccccc1)NC(=O)O[C@H]1CO[C@H]2OCC[C@@H]12
InChI Key InChIKey=BINXAIIXOUQUKC-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 9236
Found 4 hits for monomerid = 9236
TargetProtease(Human immunodeficiency virus type 1)
Kumamoto University Graduate School of Medical and Pharmaceutical Sciences
Curated by ChEMBL
Kumamoto University Graduate School of Medical and Pharmaceutical Sciences
Curated by ChEMBL
Affinity DataKi: 0.0100nMAssay Description:Inhibition of HIV1 protease dimerization in MT2 cellsMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0140nM ΔG°: -14.8kcal/mole EC50: 1.20nMpH: 6.4 T: 2°CAssay Description:The Ki values were determined by substrate cleavage assay using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-Arg-NH2. A standar...More data for this Ligand-Target Pair
TargetProtease(Human immunodeficiency virus type 1)
Kumamoto University Graduate School of Medical and Pharmaceutical Sciences
Curated by ChEMBL
Kumamoto University Graduate School of Medical and Pharmaceutical Sciences
Curated by ChEMBL
Affinity DataKi: 0.0140nMAssay Description:Inhibition of HIV1 proteaseMore data for this Ligand-Target Pair
TargetDimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A](Human immunodeficiency virus type 1)
Purdue University
Purdue University
Affinity DataKi: 0.0150nM ΔG°: -14.8kcal/molepH: 6.4 T: 2°CAssay Description:The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f...More data for this Ligand-Target Pair