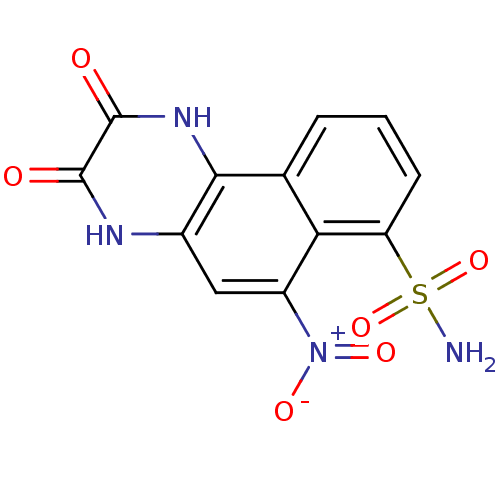
BDBM50207594 2,3-Dihydroxy-6-nitro-benzo[f]quinoxaline-7-sulfonic acid amide::2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo(f)quinoxaline::2,3-dihydroxy-6-nitro-7-sulfamoylbenzo(f)quinoxaline::3-Hydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxalin-2-ol anion::6-Nitro-2,3-dioxo-1,2,3,4,4a,10b-hexahydro-benzo[f]quinoxaline-7-sulfonic acid amide::6-Nitro-2,3-dioxo-1,2,3,4-tetrahydro-benzo[f]quinoxaline-7-sulfinic acid amide::6-Nitro-2,3-dioxo-1,2,3,4-tetrahydro-benzo[f]quinoxaline-7-sulfonic acid amide::6-Nitro-2,3-dioxo-2,3-dihydro-benzo[f]quinoxaline-7-sulfonic acid amide::6-nitro-2,3-dioxo-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide::CHEMBL222519
SMILES NS(=O)(=O)c1cccc2c1c(cc1[nH]c(=O)c(=O)[nH]c21)[N+]([O-])=O
InChI Key InChIKey=UQNAFPHGVPVTAL-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 28 hits for monomerid = 50207594
Found 28 hits for monomerid = 50207594
 3D Structure (crystal)
3D Structure (crystal)