BDBM205478 PDD00013907 (1)
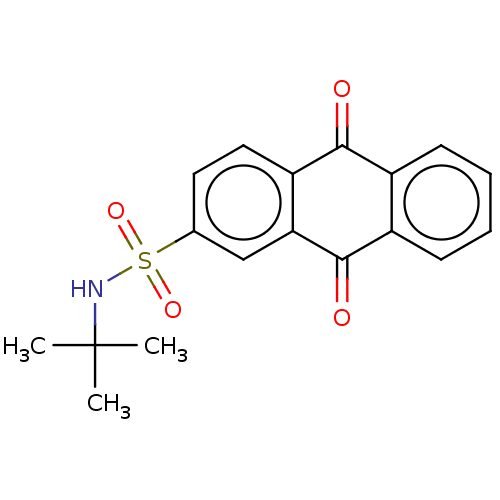
SMILES CC(C)(C)NS(=O)(=O)c1ccc2c(c1)C(=O)c3ccccc3C2=O
InChI Key InChIKey=LWNJBYGFPRLQHH-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 4 hits for monomerid = 205478
Found 4 hits for monomerid = 205478
Affinity DataKd: 1.5nMAssay Description:Binding affinity to human PARG assessed as dissociation constant by SPR analysisMore data for this Ligand-Target Pair
Affinity DataEC50: 3.00E+4nMAssay Description:Inhibition of PARG in human HeLa cells assessed as induction of MMS-induced PAR chains preincubated for 1 hr followed by MMS addition and measured af...More data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+4nMpH: 7.4Assay Description:Briefly, PARG in vitro assays were conducted in a total volume of 15 μL in a standard 384-well format. A total of 5 μL of human full length...More data for this Ligand-Target Pair
Affinity DataEC50: 8.10E+4nMAssay Description:Inhibition of human full length PARG using Bt-NAD ribosylated PARP1 substrate after 10 mins by TR-FRET assayMore data for this Ligand-Target Pair
 3D Structure (crystal)
3D Structure (crystal)