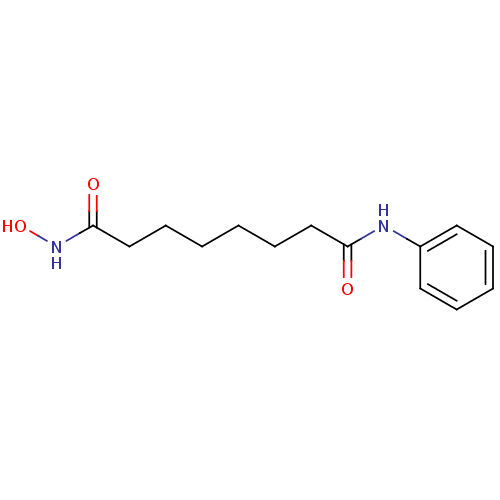
BDBM19149 CHEMBL98::N-hydroxy-N'-phenyloctanediamide::SAHA::US10011611, SAHA::US10188756, Compound SAHA::US11207431, SAHA::US11505523, Compound SAHA::US20240327418, Example SAHA::US9115116, SAHA::US9353061, SAHA::US9428447, SAHA::US9695181, Vorinostat::Vorinostat::Zolinza::cid_5311::suberoylanilide hydroxamic acid
SMILES c1ccc(cc1)NC(=O)CCCCCCC(=O)NO
InChI Key InChIKey=WAEXFXRVDQXREF-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 2062 hits for monomerid = 19149
Found 2062 hits for monomerid = 19149
Affinity DataIC50: 0.230nMT: 2°CAssay Description:Enzymatic activity was tested in 96-well or 384-well flat microwell plate by fluorescence detection and taking Ac-Lys-Tyr-Lys(Ac)-AMC as substrate. T...More data for this Ligand-Target Pair
TargetPlatelet-derived growth factor receptor alpha/beta(Mouse)
China Pharmaceutical University
Curated by ChEMBL
China Pharmaceutical University
Curated by ChEMBL
Affinity DataIC50: 0.350nMAssay Description:Inhibition of HDAC in human HeLa cells nuclear extract incubated for 30 mins by multiplate reader analysisMore data for this Ligand-Target Pair
Affinity DataKi: 0.600nM ΔG°: -12.6kcal/mole IC50: 1.10nMT: 2°CAssay Description:The fluorescence anisotropy assay was performed with fluorescein-labeled SAHA (fl-SAHA, gift of C. Fierke, University of Michigan) and measured using...More data for this Ligand-Target Pair
Affinity DataIC50: 0.850nMAssay Description:Inhibition of HDAC1 in human HeLa cells nuclear extract incubated for 30 mins by multiplate reader analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 1nMAssay Description:Inhibition of HDAC9 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 1nMAssay Description:Inhibition of HDAC4 (unknown origin)More data for this Ligand-Target Pair
Affinity DataKi: 1nMAssay Description:Displacement of fluorescent 5-(3-(3-(4-((4-((7-(hydroxyamino)-7-oxoheptyl)carbamoyl)phenylamino)methyl)-1H-1,2,3-triazol-1-yl)propyl)thioureido)-2-(3...More data for this Ligand-Target Pair
Affinity DataIC50: 1nMAssay Description:Inhibition of HDAC5 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 1nMAssay Description:Inhibition of HDAC6 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 1.40nMAssay Description:Inhibition of human KDAC3More data for this Ligand-Target Pair
Affinity DataIC50: 1.40nMAssay Description:Inhibition of human KDAC6More data for this Ligand-Target Pair
Affinity DataIC50: 1.70nMAssay Description:Inhibition of human recombinant HDAC2 after 30 mins by fluorometric assayMore data for this Ligand-Target Pair
Affinity DataKi: 1.70nMAssay Description:Binding affinity to recombinant human HDAC3 assessed as inhibition constantMore data for this Ligand-Target Pair
Affinity DataKi: 1.90nMAssay Description:Binding affinity to recombinant human HDAC1 assessed as inhibition constantMore data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:Inhibition of recombinant human HDAC2More data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:All histone deacetylases were purchased from BPS Bioscience. The substrates, Broad Substrate A, and Broad Substrate B, were synthesized and are now a...More data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:Inhibition of recombinant human HDAC1More data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:Inhibition of recombinant human HDAC6More data for this Ligand-Target Pair
Affinity DataIC50: 2nMpH: 7.4 T: 2°CAssay Description:Purified HDAC1-9 (0.5~5 nM) were incubated with 2 μM carboxyfluorescein (FAM)-labeled acetylated peptide substrate A or B and test compound for ...More data for this Ligand-Target Pair
TargetHistone deacetylase 3/Nuclear receptor corepressor 2 (HDAC3/NCoR2)(Human)
The Broad Institute of Harvard and Mit
Curated by ChEMBL
The Broad Institute of Harvard and Mit
Curated by ChEMBL
Affinity DataKi: 2nMAssay Description:Displacement of fluorescent 5-(3-(3-(4-((4-((7-(hydroxyamino)-7-oxoheptyl)carbamoyl)phenylamino)methyl)-1H-1,2,3-triazol-1-yl)propyl)thioureido)-2-(3...More data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:Inhibition of recombinant human HDAC3More data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:Inhibition of HDAC2 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 2nMT: 2°CAssay Description:Inhibition of HDAC6 (unknown origin) assessed as fluorescence intensity measured after 60 mins incubation at room temperature by trypsin-free microfl...More data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:Inhibition of HDAC3 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 2nMAssay Description:Inhibition of HDAC7 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 2.80nMAssay Description:Inhibition of recombinant human full length HDAC6 expressed in baculovirus infected Sf9 insect cells using p53 (379 to 382 residues) derived RHKKAc a...More data for this Ligand-Target Pair
Affinity DataIC50: 3nMT: 2°CAssay Description:Inhibition of HDAC3 (unknown origin) assessed as fluorescence intensity measured after 60 mins incubation at room temperature by trypsin-free microfl...More data for this Ligand-Target Pair
TargetHistone deacetylase 1/REST corepressor 3(Human)
Alma Mater Studiorum-University of Bologna
Curated by ChEMBL
Alma Mater Studiorum-University of Bologna
Curated by ChEMBL
Affinity DataKi: 3nMAssay Description:Inhibition of HDAC1/CoREST3 in HEK293 whole cell extract using fluorescent acetylated histone peptide as substrate after 60 mins by fluorescence base...More data for this Ligand-Target Pair
Affinity DataIC50: 3nMAssay Description:Inhibition of HDAC1 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 3nMAssay Description:Inhibition of human recombinant HDAC1 using Boc-Lys(Ac)-AMC as substrate incubated for 1 hrs in dark by fluorescence based assayMore data for this Ligand-Target Pair
Affinity DataIC50: 3nMpH: 7.4 T: 2°CAssay Description:Purified HDAC1-9 (0.5~5 nM) were incubated with 2 μM carboxyfluorescein (FAM)-labeled acetylated peptide substrate A or B and test compound for ...More data for this Ligand-Target Pair
Affinity DataIC50: 3nMAssay Description:All histone deacetylases were purchased from BPS Bioscience. The substrates, Broad Substrate A, and Broad Substrate B, were synthesized and are now a...More data for this Ligand-Target Pair
TargetPlatelet-derived growth factor receptor alpha/beta(Mouse)
China Pharmaceutical University
Curated by ChEMBL
China Pharmaceutical University
Curated by ChEMBL
Affinity DataIC50: 3.10nMAssay Description:Inhibition of human HDAC3/NCOR2 using RHKKAc as substrate by fluorescence based assayMore data for this Ligand-Target Pair
Affinity DataIC50: 3.10nMAssay Description:Inhibition of HDAC3 (unknown origin) using fluorogenic peptide RHKKAc-AMC as substrate by fluorescence-based assayMore data for this Ligand-Target Pair
Affinity DataIC50: 3.10nMAssay Description:Inhibition of HDAC3 (unknown origin) using Fluor de lys as substrate by fluorescence based analysisMore data for this Ligand-Target Pair
Affinity DataIC50: 3.30nMAssay Description:Inhibition of HDAC3 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 3.30nMAssay Description:Inhibition of HDAC3 (unknown origin) expressed in baculovirus infected in Sf9 cells using RHKKAc as substrate preincubated for 5 to 10 mins followed ...More data for this Ligand-Target Pair
Affinity DataIC50: 3.90nMAssay Description:Inhibition of full length human N-terminal GST-tagged HDAC6 using fluorogenic acetylated peptide as substrate incubated for 30 mins by fluorescence p...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMT: 2°CAssay Description:Inhibition of HDAC1 (unknown origin) assessed as fluorescence intensity measured after 60 mins incubation at room temperature by trypsin-free microfl...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of HDAC1 (unknown origin) using Fluor de Lys substrate by fluorescence assay relative to controlMore data for this Ligand-Target Pair
Affinity DataIC50: 4nMpH: 7.4 T: 2°CAssay Description:Purified HDAC1-9 (0.5~5 nM) were incubated with 2 μM carboxyfluorescein (FAM)-labeled acetylated peptide substrate A or B and test compound for ...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:All histone deacetylases were purchased from BPS Bioscience. The substrates, Broad Substrate A, and Broad Substrate B, were synthesized and are now a...More data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of HDAC3 (unknown origin) after 60 mins by fluorescence assayMore data for this Ligand-Target Pair
Affinity DataIC50: 4nMAssay Description:Inhibition of HDAC11 (unknown origin)More data for this Ligand-Target Pair
Affinity DataIC50: 4.20nMAssay Description:Inhibition of HDAC1 (unknown origin) expressed in baculovirus infected in Sf9 cells using RHKKAc as substrate preincubated for 5 to 10 mins followed ...More data for this Ligand-Target Pair
Affinity DataIC50: 4.20nMAssay Description:Inhibition of HDAC1 (unknown origin)More data for this Ligand-Target Pair
Activity Spreadsheet -- ITC Data from BindingDB
 Found 2 hits for monomerid = 19149
Found 2 hits for monomerid = 19149
ITC DataΔG°: -8.36kcal/mole −TΔS°: 2.42kcal/mole ΔH°: -10.9kcal/mole logk: 1.36E+6
T: 25.00°C
T: 25.00°C
ITC DataΔG°: -8.36kcal/mole −TΔS°: 0.0519kcal/mole ΔH°: -8.96kcal/mole logk: 2.21E+6
pH: 7.5 T: -248.15°C
pH: 7.5 T: -248.15°C
 3D Structure (crystal)
3D Structure (crystal)