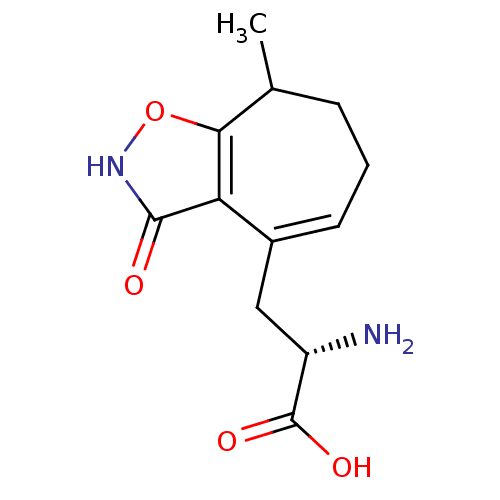
BDBM50295042 (S)-2-Amino-3-((RS)-3-hydroxy-8-methyl-7,8-dihydro-6H-cyclohepta-[d]isoxazol-4-yl)propionic acid::CHEMBL553028
SMILES CC1CCC=C(C[C@H](N)C(O)=O)c2c1o[nH]c2=O
InChI Key InChIKey=OGDYFTNOLZTFCD-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 16 hits for monomerid = 50295042
Found 16 hits for monomerid = 50295042
Affinity DataEC50: 7.00E+4nMAssay Description:Agonist activity at rat recombinant GluR1 flip isoform expressed in HEK293 cells assessed as increase in intracellular calcium level by Fluo-4/AM ass...More data for this Ligand-Target Pair
Affinity DataEC50: 1.60E+5nMAssay Description:Agonist activity at rat recombinant GluR2(Q) RNA-edited isoform expressed in HEK293 cells assessed as increase in intracellular calcium level by Fluo...More data for this Ligand-Target Pair
Affinity DataEC50: 4.80E+4nMAssay Description:Agonist activity at rat recombinant GluR3 flip isomer expressed in HEK293 cells assessed as increase in intracellular calcium level by Fluo-4/AM assa...More data for this Ligand-Target Pair
Affinity DataEC50: 3.50E+4nMAssay Description:Agonist activity at rat recombinant GluR4 flip isomer expressed in HEK293 cells assessed as increase in intracellular calcium level by Fluo-4/AM assa...More data for this Ligand-Target Pair
Affinity DataEC50: 860nMAssay Description:Agonist activity at rat recombinant GluR5(Q) RNA-edited isoform expressed in HEK293 cells assessed as increase in intracellular calcium level by Fluo...More data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+6nMAssay Description:Agonist activity at rat recombinant GluR6(Q) RNA-edited isoform expressed in HEK293 cells assessed as increase in intracellular calcium level by Fluo...More data for this Ligand-Target Pair
Affinity DataEC50: 660nMAssay Description:Agonist activity at ConA-treated rat GluR5(Q) RNA-edited isoform expressed in Xenopus laevis oocytes assessed as glutamic acid-induced maximal curren...More data for this Ligand-Target Pair
Affinity DataEC50: 6.20E+4nMAssay Description:Agonist activity at ConA-treated rat GluR1 flip isoform expressed in Xenopus laevis oocytes assessed as glutamic acid-induced maximal current by two-...More data for this Ligand-Target Pair
Affinity DataEC50: >1.00E+6nMAssay Description:Agonist activity at ConA-treated rat NR1/NR2A expressed in Xenopus laevis oocytes assessed as glycine-induced maximal current by two-electrode voltag...More data for this Ligand-Target Pair
Affinity DataKi: 5.08nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR5(Q) RNA-edited isoform expressed in baculovirus infected Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.11E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR3 flop isoform expressed in baculovirus infected Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.38E+3nMAssay Description:Displacement of [3H]SYM from rat recombinant GluR7A expressed in baculovirus infected Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 1.55E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR1 flop isoform expressed in baculovirus infected Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 2.20E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR4 flop isoform expressed in baculovirus infected Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: 6.31E+3nMAssay Description:Displacement of [3H]AMPA from rat recombinant GluR2(R) RNA-edited flop isoform expressed in baculovirus infected Sf9 cellsMore data for this Ligand-Target Pair
Affinity DataKi: >1.00E+5nMAssay Description:Displacement of [3H]KA from rat recombinant GluR6(VCR) RNA-edited isoform expressed in baculovirus infected Sf9 cellsMore data for this Ligand-Target Pair