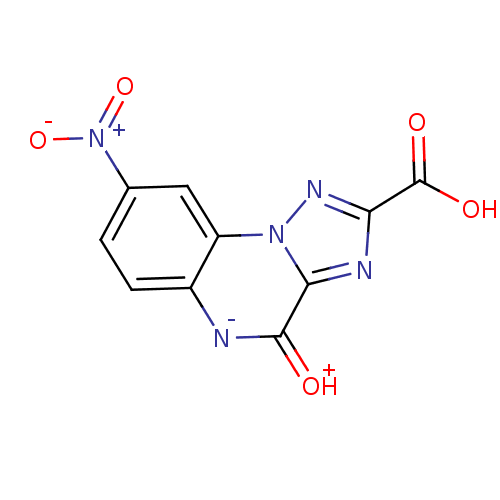
BDBM50078511 8-Nitro-4-oxo-4,5-dihydro-[1,2,4]triazolo[1,5-a]quinoxaline-2-carboxylic acid::CHEMBL311860
SMILES OC(=O)c1nc2n(n1)c1cc(ccc1[n-]c2=[OH+])[N+]([O-])=O
InChI Key InChIKey=BFJRNXBUOBCWCO-UHFFFAOYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 50078511
Found 3 hits for monomerid = 50078511
Affinity DataEC50: >1.00E+5nMAssay Description:Functional antagonism at the N-methyl-D-aspartate glutamate receptor 1 was demonstarted by the ability to inhibit the binding of the channel-blocking...More data for this Ligand-Target Pair
Affinity DataKi: 7.94E+4nMAssay Description:Binding affinity towards N-methyl-D-aspartate glutamate receptor 1 in rat cortical membranes using [3H]glycine as radioligandMore data for this Ligand-Target Pair
Affinity DataKi: 7.94E+4nMAssay Description:Binding selectivity towards N-methyl-D-aspartate glutamate receptor 1 was determined by using [3H]- glycine/NMDAMore data for this Ligand-Target Pair