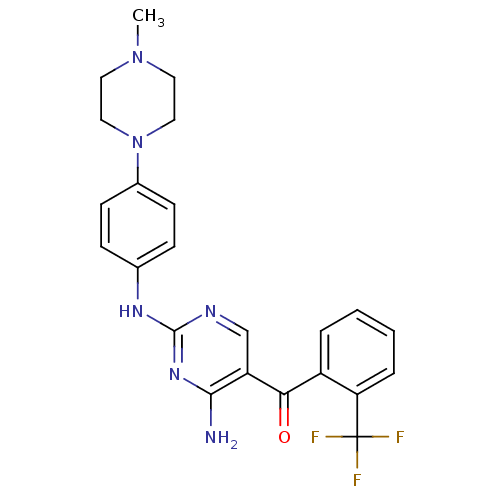
BDBM12602 2,4-Diamino-5-ketopyrimidine 20::2-N-[4-(4-methylpiperazin-1-yl)phenyl]-5-{[2-(trifluoromethyl)phenyl]carbonyl}pyrimidine-2,4-diamine
SMILES CN1CCN(CC1)c1ccc(Nc2ncc(C(=O)c3ccccc3C(F)(F)F)c(N)n2)cc1
InChI Key InChIKey=DUOCMHSNSNZKRJ-UHFFFAOYSA-N
Data 3 KI
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 12602
Found 3 hits for monomerid = 12602
Affinity DataKi: 821nMAssay Description:Enzymes were assayed with retinoblastoma substrate in 384-well plates containing diluted test compounds. Final ATP concentration was 3x the respectiv...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nM ΔG°: >-7.09kcal/molepH: 7.0 T: 2°CAssay Description:Enzymes were assayed with retinoblastoma substrate in 384-well plates containing diluted test compounds. Final ATP concentration was 3x the respectiv...More data for this Ligand-Target Pair
Affinity DataKi: >1.00E+4nMAssay Description:Enzymes were assayed with retinoblastoma substrate in 384-well plates containing diluted test compounds. Final ATP concentration was 3x the respectiv...More data for this Ligand-Target Pair