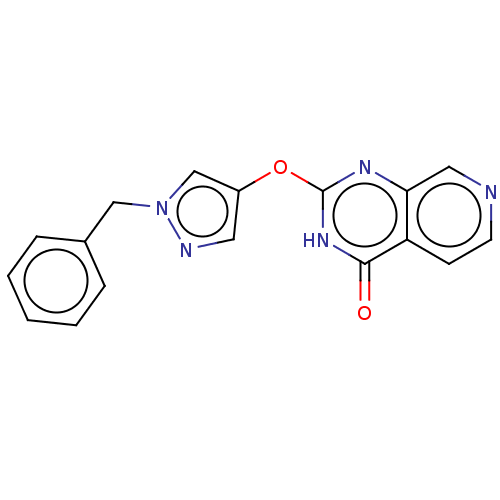
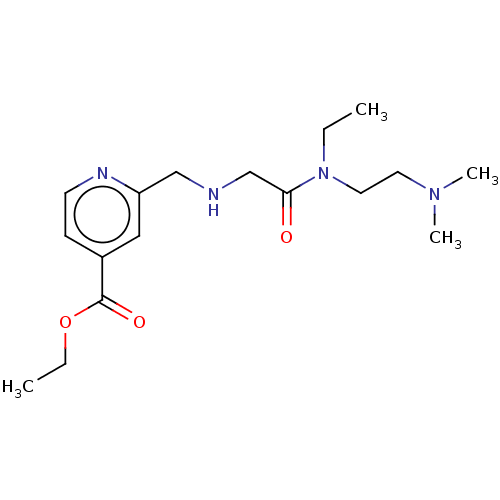
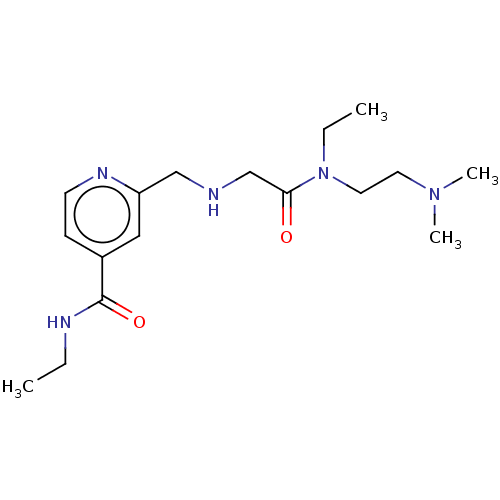
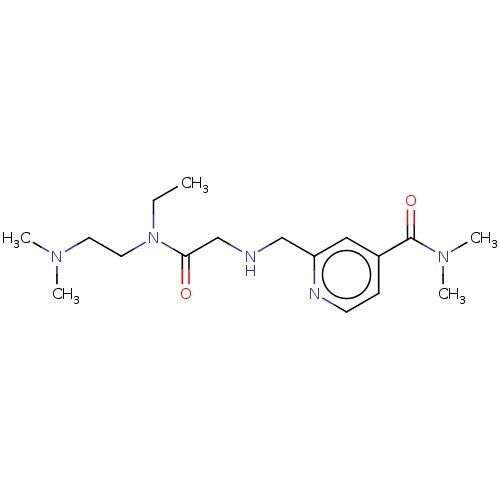
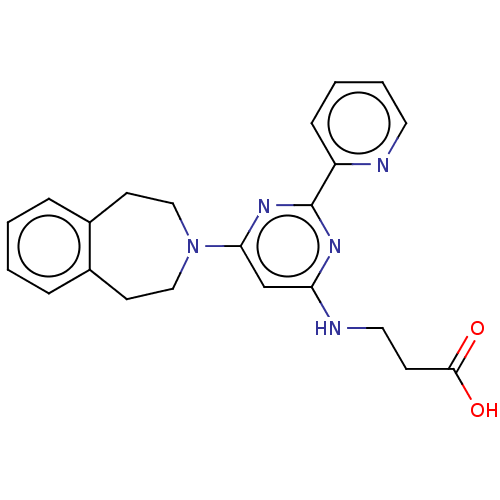
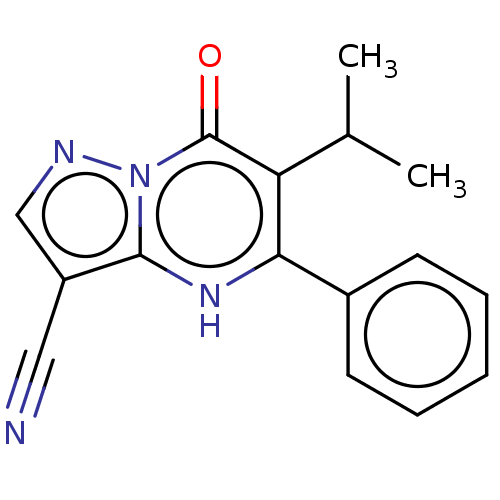
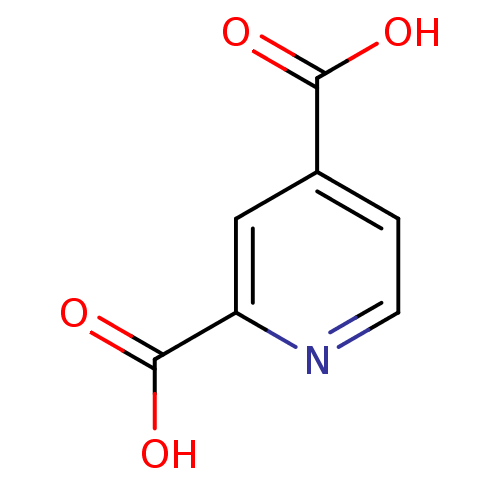
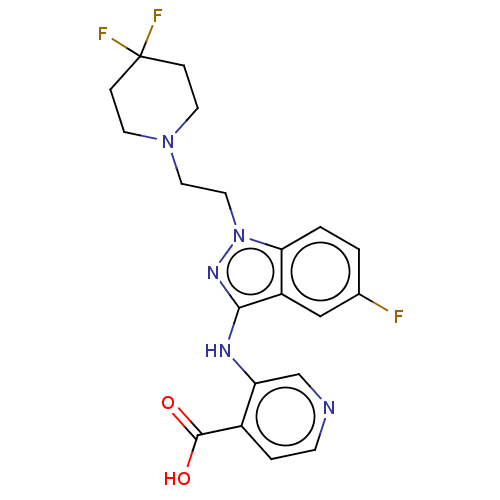
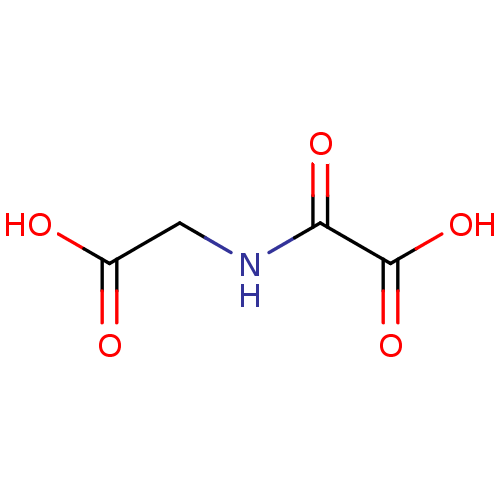
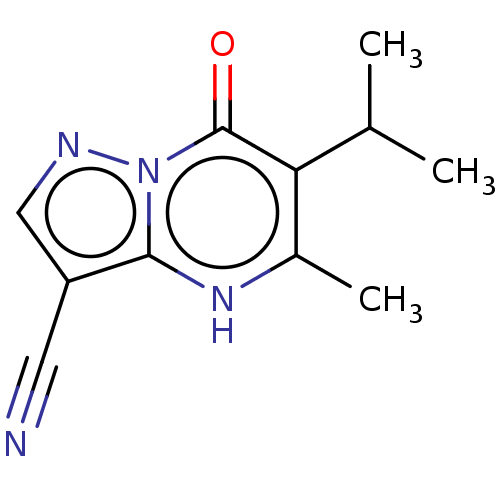
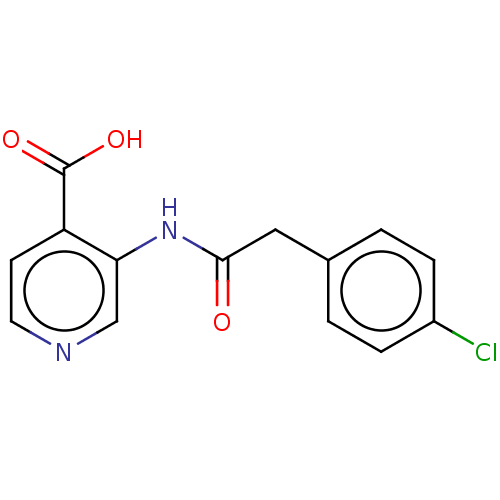
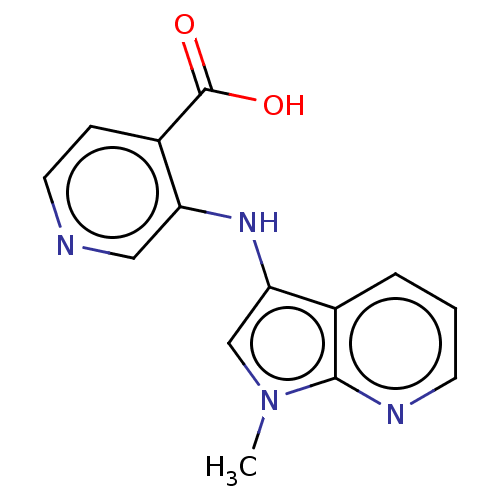
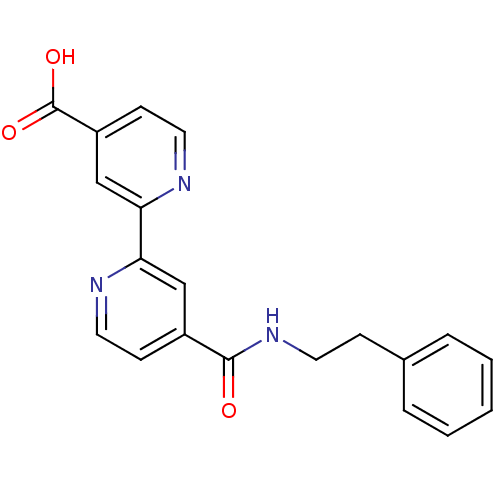
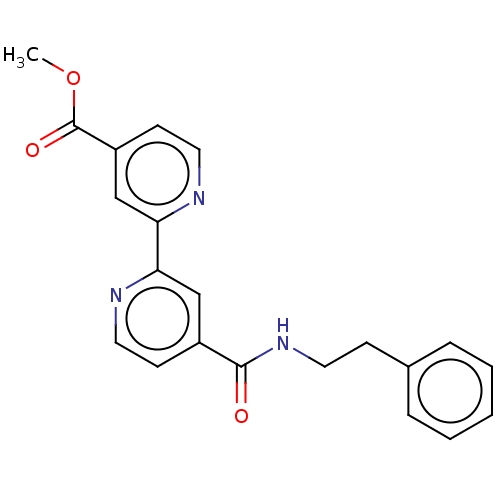
Affinity DataIC50: 15nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 20nMAssay Description:We previously described 4-carboxy-2-triazolopyridines as selective KDM2 inhibitors (England et al., 2014). In efforts to find alternative scaffolds b...More data for this Ligand-Target Pair
Affinity DataIC50: 27nMAssay Description:Inhibition of KDM5D (unknown origin) in presence of 4 uM 2-OGMore data for this Ligand-Target Pair
Affinity DataIC50: 60nMAssay Description:We previously described 4-carboxy-2-triazolopyridines as selective KDM2 inhibitors (England et al., 2014). In efforts to find alternative scaffolds b...More data for this Ligand-Target Pair
Affinity DataIC50: 69nMAssay Description:We previously described 4-carboxy-2-triazolopyridines as selective KDM2 inhibitors (England et al., 2014). In efforts to find alternative scaffolds b...More data for this Ligand-Target Pair
Affinity DataIC50: 77nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 100nMAssay Description:We previously described 4-carboxy-2-triazolopyridines as selective KDM2 inhibitors (England et al., 2014). In efforts to find alternative scaffolds b...More data for this Ligand-Target Pair
Affinity DataIC50: 200nMAssay Description:We previously described 4-carboxy-2-triazolopyridines as selective KDM2 inhibitors (England et al., 2014). In efforts to find alternative scaffolds b...More data for this Ligand-Target Pair
Affinity DataIC50: 558nMAssay Description:Inhibition of KDM5D (unknown origin) in presence of 300 uM 2-OGMore data for this Ligand-Target Pair
Affinity DataIC50: 1.00E+3nMAssay Description:We previously described 4-carboxy-2-triazolopyridines as selective KDM2 inhibitors (England et al., 2014). In efforts to find alternative scaffolds b...More data for this Ligand-Target Pair
Affinity DataIC50: 1.50E+3nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+3nMAssay Description:Inhibition of KDM5D (unknown origin ) by HTMS assayMore data for this Ligand-Target Pair
Affinity DataIC50: 2.50E+3nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 2.70E+3nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 5.80E+3nMpH: 7.5Assay Description:The demethylase AlphaScreen assay was performed in 384-well plate format using white proxiplates (PerkinElmer), and transfer of compound (100 nl) was...More data for this Ligand-Target Pair
Affinity DataIC50: 6.00E+3nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 1.10E+4nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 1.40E+4nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 1.80E+4nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 2.20E+4nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 5.30E+4nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 2.00E+5nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 2.90E+5nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
Affinity DataIC50: 4.70E+5nMT: 2°CAssay Description:For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti...More data for this Ligand-Target Pair
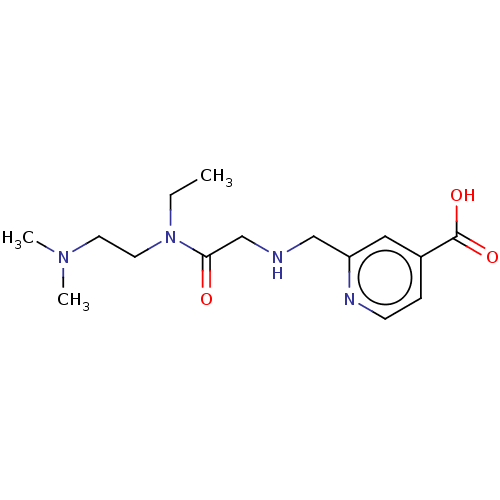

 3D Structure (crystal)
3D Structure (crystal)