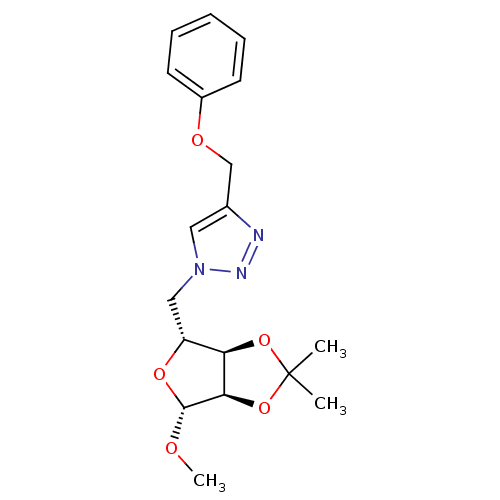
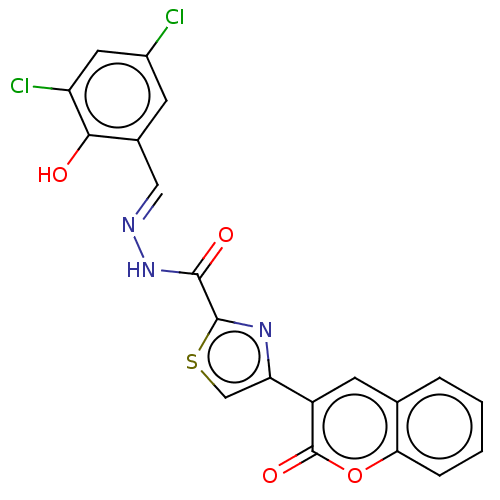
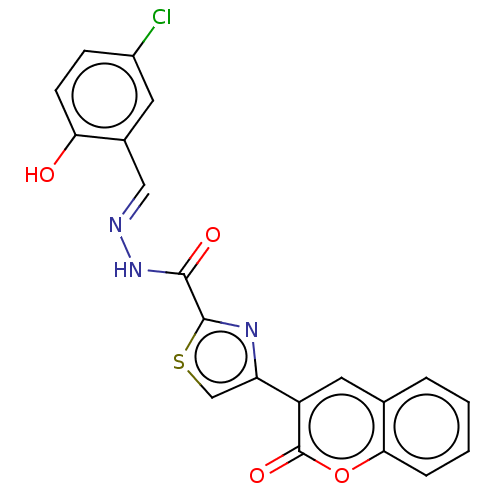
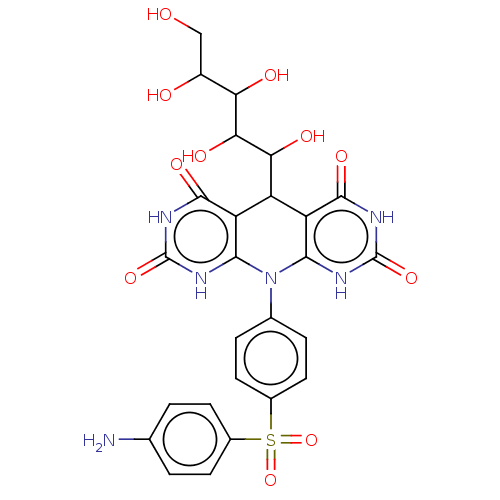
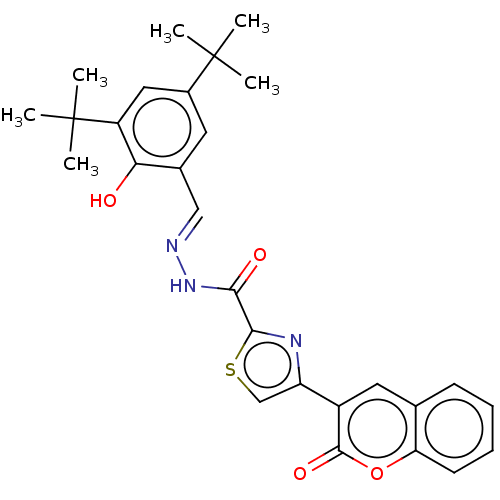
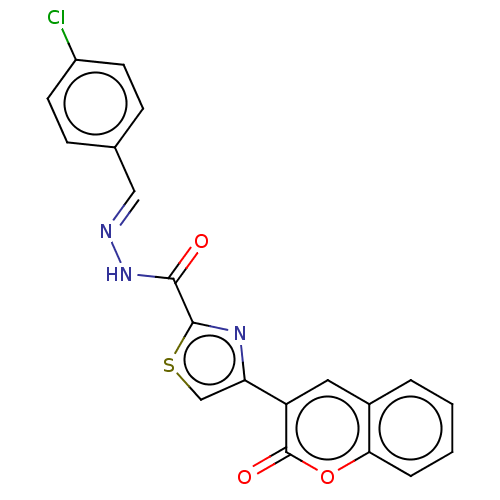
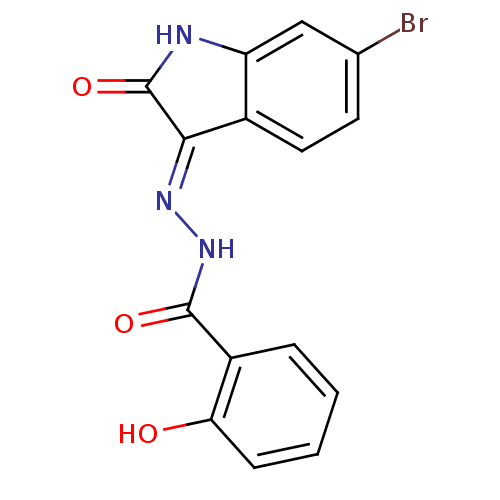
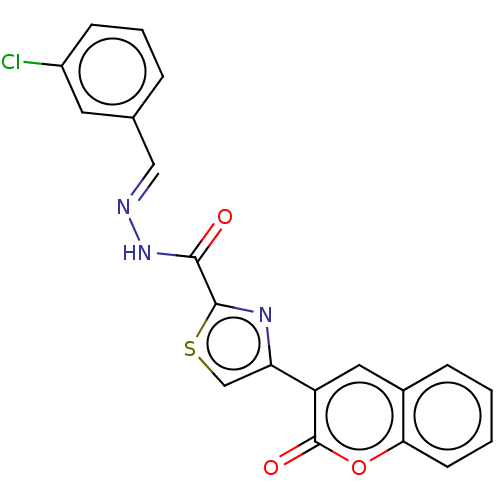
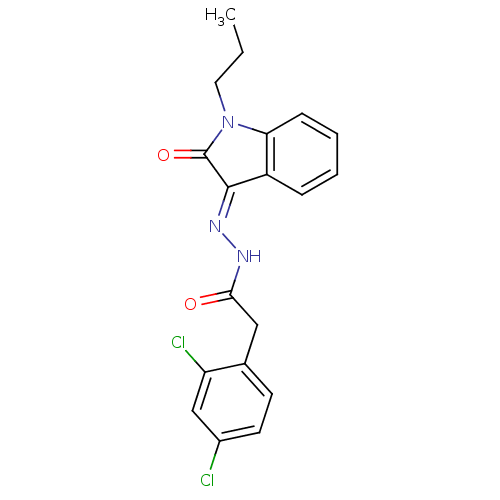
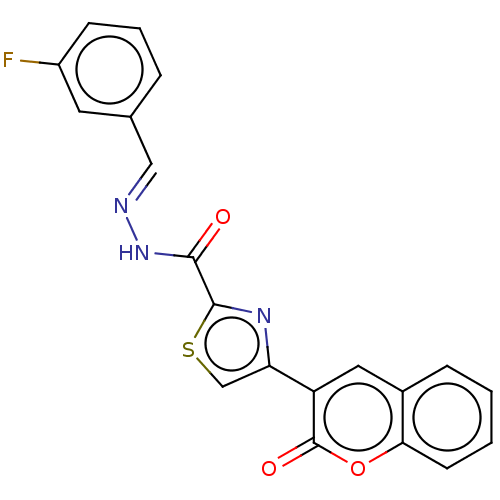
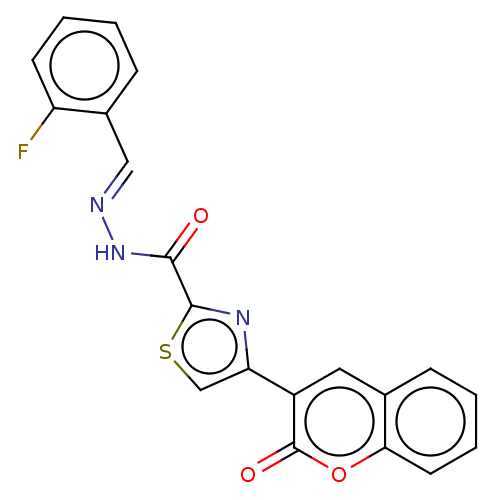
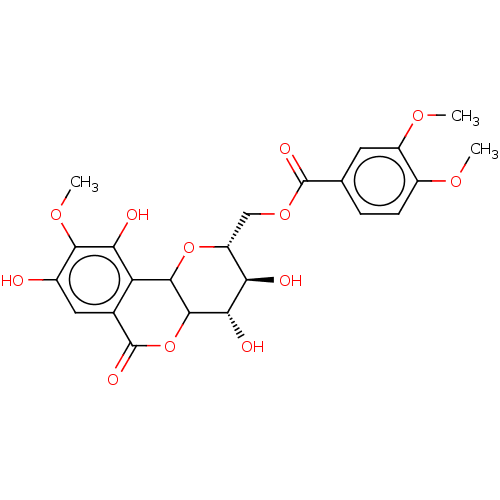
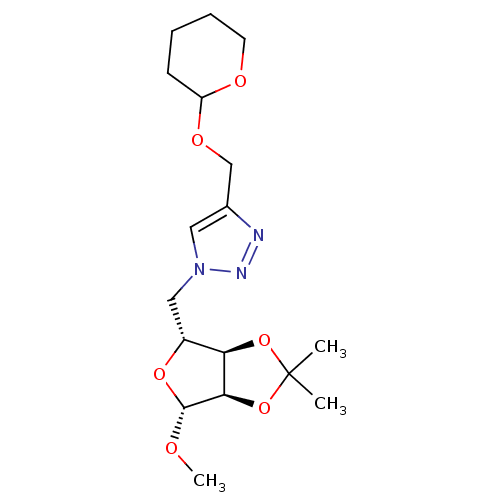
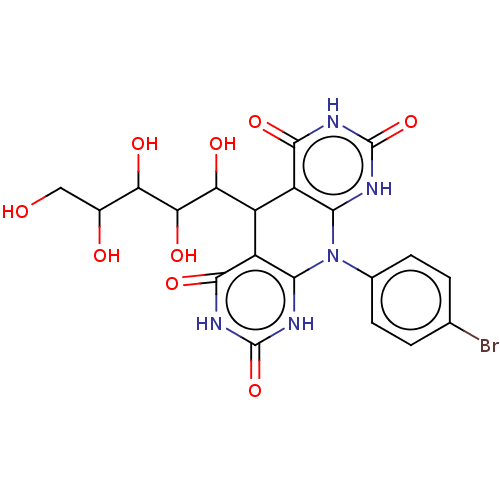
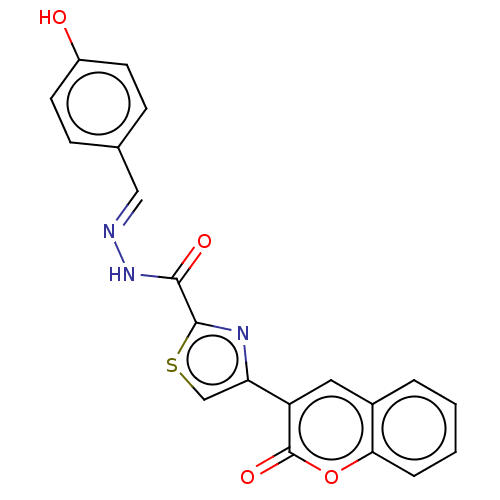
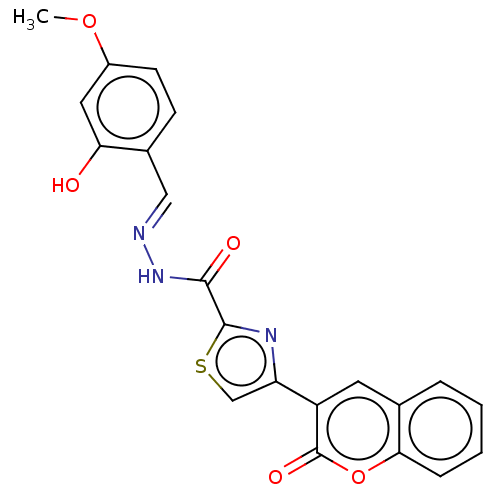
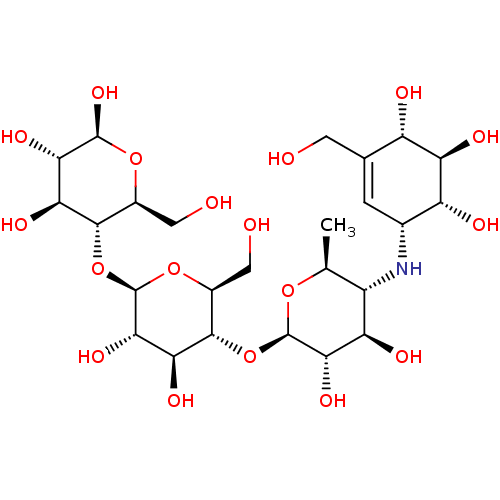
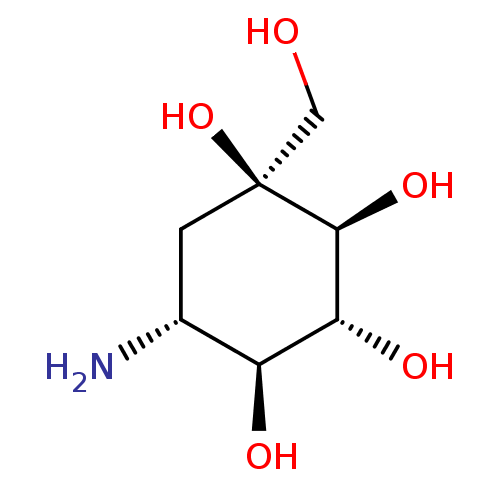
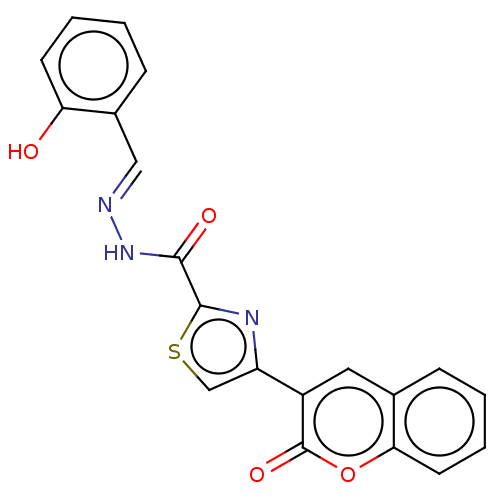
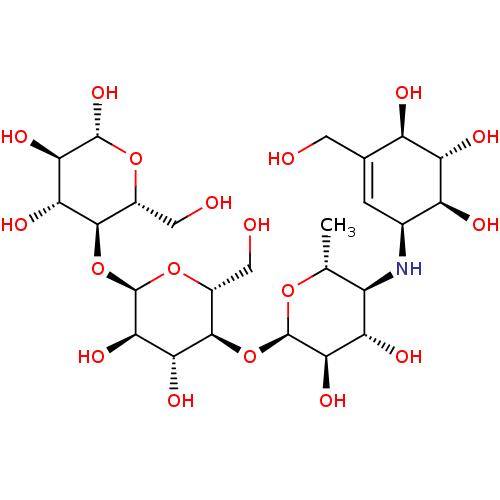
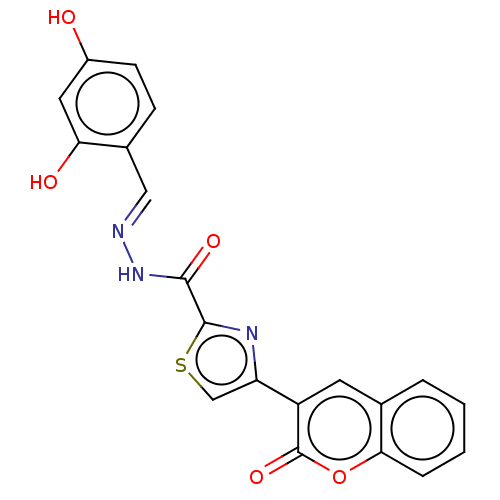
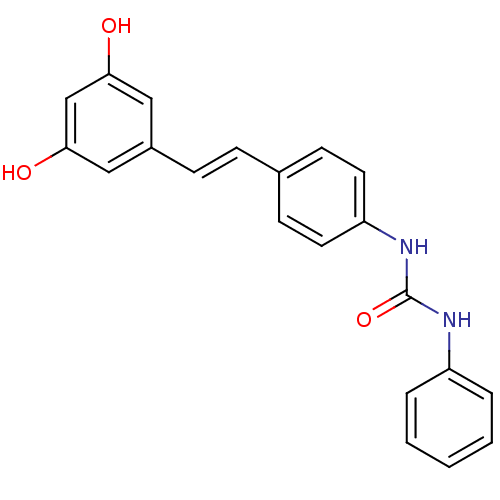
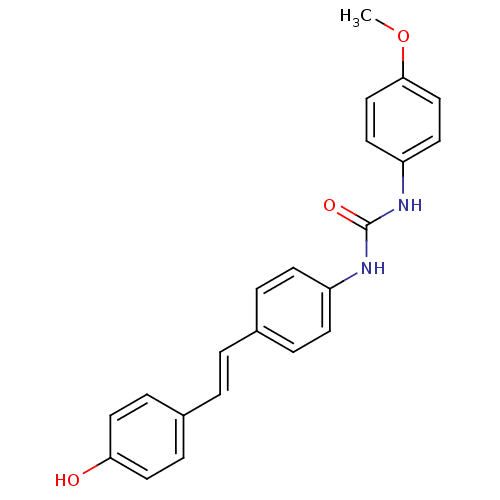
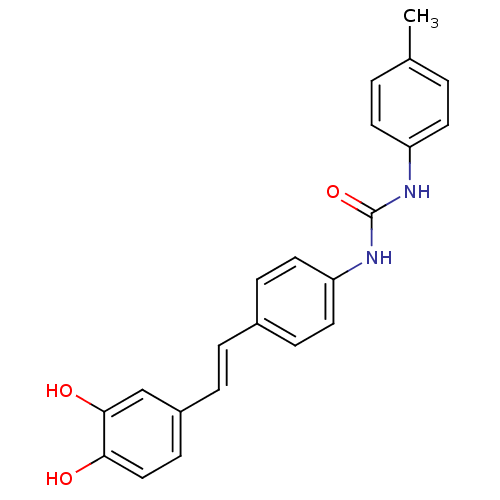
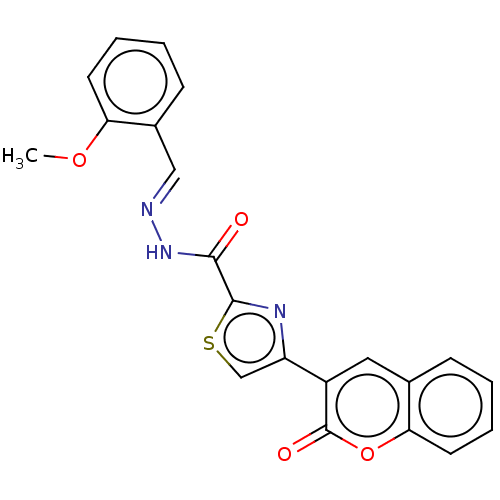
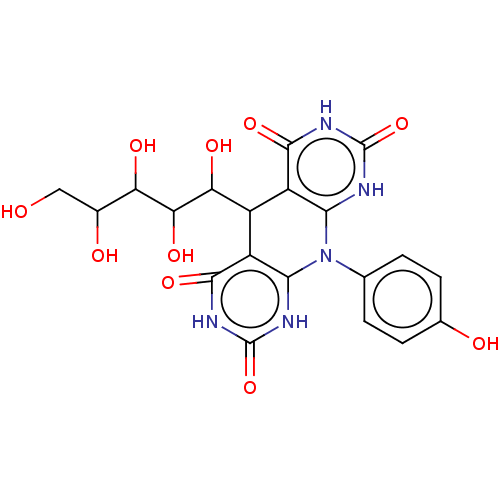
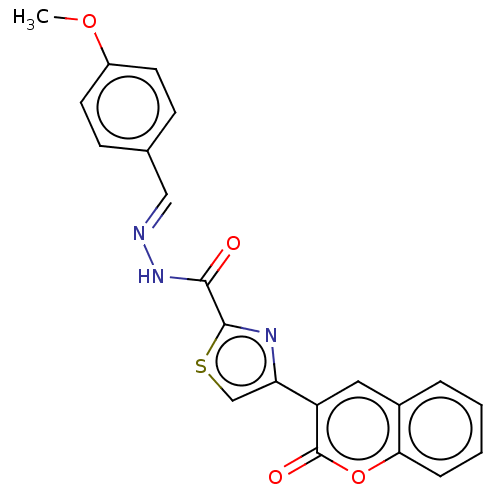
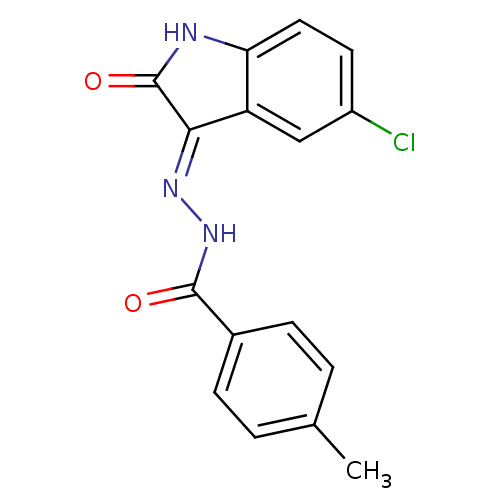
Affinity DataIC50: 2.20E+3nMAssay Description:10 µL of test samples (5 mg/mL DMSO solution) were reconstituted in 100 µL of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 3.00E+3nMAssay Description:10 µL of test samples (5 mg/mL DMSO solution) were reconstituted in 100 µL of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 3.30E+3nMAssay Description:10 µL of test samples (5 mg/mL DMSO solution) were reconstituted in 100 µL of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 3.80E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 5.20E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 5.70E+3nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 6.24E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 8.23E+3nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 9.60E+3nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataIC50: 1.02E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 1.06E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 1.18E+4nMAssay Description:10 µL of test samples (5 mg/mL DMSO solution) were reconstituted in 100 µL of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 1.20E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 1.40E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 1.49E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 1.51E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 1.60E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 1.66E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 1.83E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 1.83E+4nMAssay Description:10 µL of test samples (5 mg/mL DMSO solution) were reconstituted in 100 µL of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 2.26E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 2.36E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 2.46E+4nMpH: 7.0 T: 2°CAssay Description:α-glucosidase (25 μL, 0.2 U/mL), 25 μL of various concentrations of samples, and 175 μL of 50 mM sodium phosphate buffer (pH 7.0)...More data for this Ligand-Target Pair
Affinity DataIC50: 2.47E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair
Affinity DataIC50: 2.51E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataIC50: 2.52E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 2.99E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 3.39E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 using 4-nitrophenyl-alpha-D-glucopyranoside as substrate preincubated for 30 mins followed by substrate a...More data for this Ligand-Target Pair
Affinity DataIC50: 3.60E+4nMAssay Description:Inhibition of baker's yeast alpha-glucosidaseMore data for this Ligand-Target Pair
Affinity DataIC50: 3.72E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 3.83E+4nMpH: 6.8 T: 2°CAssay Description:Reaction mixture (100 µL) contained 70 µL 50 mM Na2HPO4 buffer, pH 6.8, 10 µL test compound (0.5 mM), followed by the addition of 10 &...More data for this Ligand-Target Pair
Affinity DataIC50: 3.86E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 4.33E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 4.98E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 using 4-nitrophenyl-alpha-D-glucopyranoside as substrate preincubated for 30 mins followed by substrate a...More data for this Ligand-Target Pair
Affinity DataIC50: 5.88E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataIC50: 6.01E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataIC50: 6.36E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataIC50: 6.43E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 6.43E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataIC50: 6.85E+4nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataIC50: 7.49E+4nMpH: 7.0Assay Description:In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth...More data for this Ligand-Target Pair
Affinity DataIC50: 8.17E+4nMpH: 6.8Assay Description:The test compounds were dissolved in DMSO to prepare the required distributing concentration. α-Glucosidase inhibitory activity was assayed by u...More data for this Ligand-Target Pair
Affinity DataIC50: 8.35E+4nMAssay Description:10 µL of test samples (5 mg/mL DMSO solution) were reconstituted in 100 µL of 100 mM-phosphate buffer (pH6.8) in 96-well microplate and incubated wit...More data for this Ligand-Target Pair
Affinity DataIC50: 8.51E+4nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 8.57E+4nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 using 4-nitrophenyl-alpha-D-glucopyranoside as substrate preincubated for 30 mins followed by substrate a...More data for this Ligand-Target Pair
Affinity DataIC50: 9.08E+4nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 9.82E+4nMAssay Description:The assay was performed using isolated phenolics from maize, and inhibition was determined according to previously described method.More data for this Ligand-Target Pair
Affinity DataIC50: 1.02E+5nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataIC50: 1.05E+5nMAssay Description:All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos...More data for this Ligand-Target Pair
Affinity DataIC50: 1.09E+5nMAssay Description:Inhibition of yeast alpha-glucosidase MAL12 assessed as inhibition of p-nitrophenol release by spectrophotometryMore data for this Ligand-Target Pair